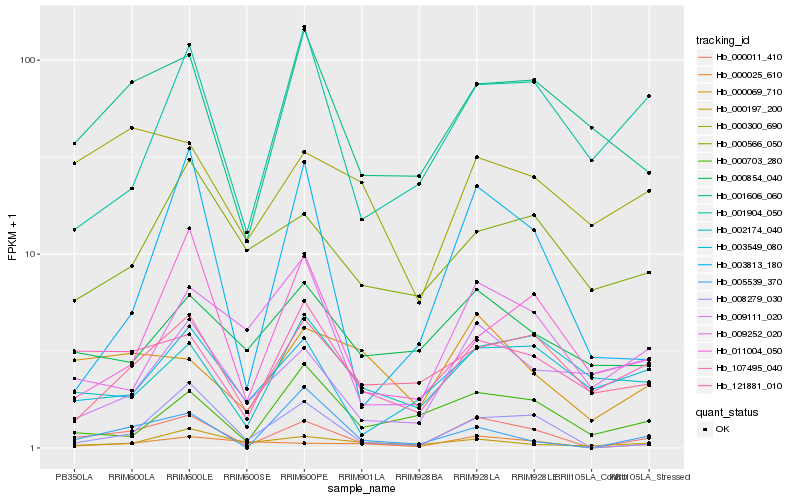
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000011_410 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008279_030 |
0.2186555092 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_107495_040 |
0.2217890837 |
- |
- |
PREDICTED: 12-oxophytodienoate reductase 2-like [Citrus sinensis] |
| 4 |
Hb_001606_060 |
0.237160614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000025_610 |
0.2406637641 |
- |
- |
annexin, putative [Ricinus communis] |
| 6 |
Hb_000069_710 |
0.2446451649 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 7 |
Hb_121881_010 |
0.2486318669 |
- |
- |
hypothetical protein POPTR_0015s15770g [Populus trichocarpa] |
| 8 |
Hb_003813_180 |
0.2543743334 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 9 |
Hb_003549_080 |
0.2550478117 |
- |
- |
PREDICTED: importin subunit alpha-4 isoform X2 [Jatropha curcas] |
| 10 |
Hb_009252_020 |
0.2572119642 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 11 |
Hb_001904_050 |
0.2631051178 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 12 |
Hb_009111_020 |
0.2646898071 |
- |
- |
PREDICTED: calcium-dependent protein kinase 20-like [Jatropha curcas] |
| 13 |
Hb_002174_040 |
0.2664477044 |
- |
- |
DNAJ heat shock family protein [Theobroma cacao] |
| 14 |
Hb_000854_040 |
0.2672084597 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 15 |
Hb_000197_200 |
0.268358846 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 16 |
Hb_011004_050 |
0.2692746393 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 17 |
Hb_000300_690 |
0.2695305583 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 18 |
Hb_000703_280 |
0.2705200025 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 19 |
Hb_000566_050 |
0.2706411314 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 20 |
Hb_005539_370 |
0.2709876719 |
- |
- |
ring finger protein, putative [Ricinus communis] |