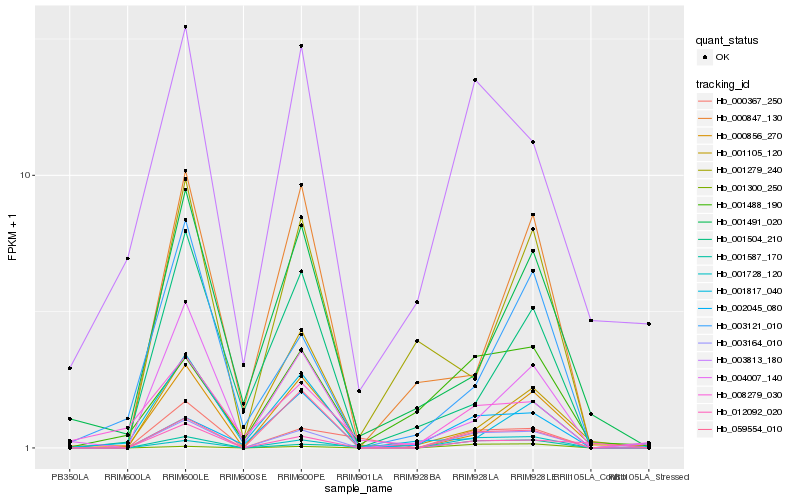
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003164_010 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_001728_120 |
0.1784426266 |
- |
- |
hypothetical protein POPTR_0001s15540g, partial [Populus trichocarpa] |
| 3 |
Hb_001587_170 |
0.2155025833 |
- |
- |
hypothetical protein B456_010G223200, partial [Gossypium raimondii] |
| 4 |
Hb_000367_250 |
0.2227573301 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_004007_140 |
0.2278762033 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002045_080 |
0.2311145026 |
- |
- |
PREDICTED: uncharacterized protein LOC105649907 [Jatropha curcas] |
| 7 |
Hb_000856_270 |
0.2347260558 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 8 |
Hb_012092_020 |
0.2359872049 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_059554_010 |
0.2521610308 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 10 |
Hb_001504_210 |
0.2628496587 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003813_180 |
0.2682527548 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 12 |
Hb_000847_130 |
0.2684041624 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 13 |
Hb_003121_010 |
0.2707951184 |
- |
- |
hypothetical protein POPTR_0018s09720g [Populus trichocarpa] |
| 14 |
Hb_001488_190 |
0.2760870813 |
- |
- |
alpha-amylase, putative [Ricinus communis] |
| 15 |
Hb_001300_250 |
0.277405608 |
- |
- |
hypothetical protein JCGZ_22020 [Jatropha curcas] |
| 16 |
Hb_001491_020 |
0.2826026536 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_001279_240 |
0.2830348764 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105633500 [Jatropha curcas] |
| 18 |
Hb_008279_030 |
0.2855711599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001817_040 |
0.2878550873 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 20 |
Hb_001105_120 |
0.2910329297 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |