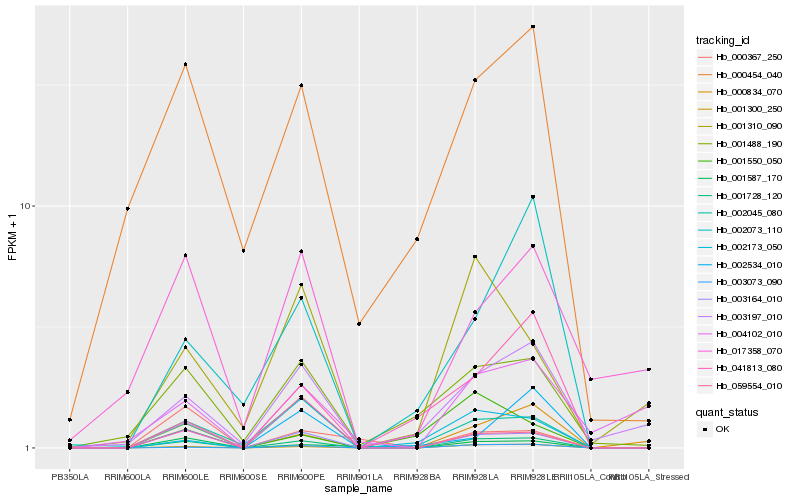
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001728_120 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s15540g, partial [Populus trichocarpa] |
| 2 |
Hb_001300_250 |
0.1161801601 |
- |
- |
hypothetical protein JCGZ_22020 [Jatropha curcas] |
| 3 |
Hb_003164_010 |
0.1784426266 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_002045_080 |
0.2074621503 |
- |
- |
PREDICTED: uncharacterized protein LOC105649907 [Jatropha curcas] |
| 5 |
Hb_003073_090 |
0.2230264732 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_0004s23920g [Populus trichocarpa] |
| 6 |
Hb_000834_070 |
0.2299005902 |
- |
- |
PREDICTED: tankyrase-1 isoform X4 [Jatropha curcas] |
| 7 |
Hb_001587_170 |
0.2500605489 |
- |
- |
hypothetical protein B456_010G223200, partial [Gossypium raimondii] |
| 8 |
Hb_001488_190 |
0.2520491408 |
- |
- |
alpha-amylase, putative [Ricinus communis] |
| 9 |
Hb_003197_010 |
0.2562156072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000454_040 |
0.2755739202 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 11 |
Hb_002173_050 |
0.2801836335 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_001310_090 |
0.2875342263 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |
| 13 |
Hb_002073_110 |
0.2899931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002534_010 |
0.2928519743 |
- |
- |
PREDICTED: expansin-A1-like [Jatropha curcas] |
| 15 |
Hb_001550_050 |
0.306814825 |
- |
- |
PREDICTED: J domain-containing protein spf31 [Eucalyptus grandis] |
| 16 |
Hb_004102_010 |
0.3070896932 |
- |
- |
F-box family protein [Theobroma cacao] |
| 17 |
Hb_059554_010 |
0.3075396325 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 18 |
Hb_017358_070 |
0.311423394 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 19 |
Hb_000367_250 |
0.3258322022 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_041813_080 |
0.3278626769 |
- |
- |
- |