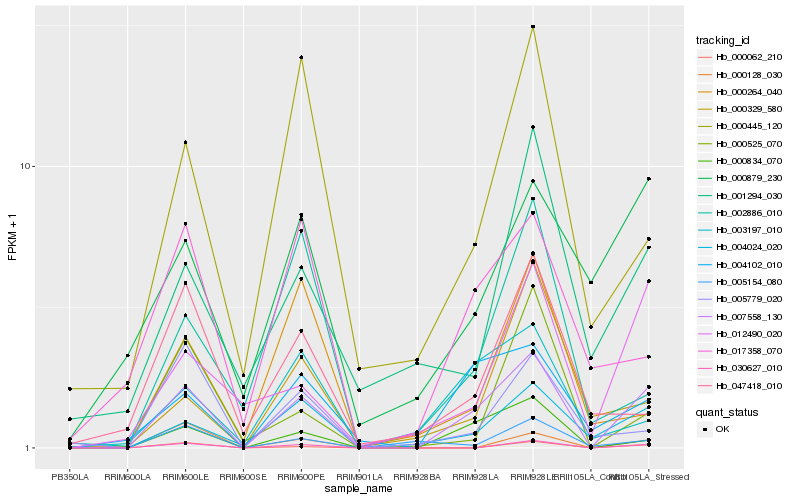
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007558_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_000834_070 |
0.2102025356 |
- |
- |
PREDICTED: tankyrase-1 isoform X4 [Jatropha curcas] |
| 3 |
Hb_004024_020 |
0.2278855337 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_030627_010 |
0.238086785 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |
| 5 |
Hb_004102_010 |
0.2504679435 |
- |
- |
F-box family protein [Theobroma cacao] |
| 6 |
Hb_000062_210 |
0.2731714765 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Pyrus x bretschneideri] |
| 7 |
Hb_012490_020 |
0.277200307 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 8 |
Hb_003197_010 |
0.2867800948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000445_120 |
0.2989256699 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_005154_080 |
0.3023523095 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640764 [Jatropha curcas] |
| 11 |
Hb_017358_070 |
0.3026289992 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 12 |
Hb_000525_070 |
0.3029849195 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like [Jatropha curcas] |
| 13 |
Hb_005779_020 |
0.3086891086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000329_580 |
0.309606793 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_047418_010 |
0.3138280147 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 16 |
Hb_000264_040 |
0.315302681 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_000879_230 |
0.3154683683 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 18 |
Hb_002886_010 |
0.3162529576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000128_030 |
0.3165374142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001294_030 |
0.3172706068 |
- |
- |
PREDICTED: uncharacterized protein LOC105644307 [Jatropha curcas] |