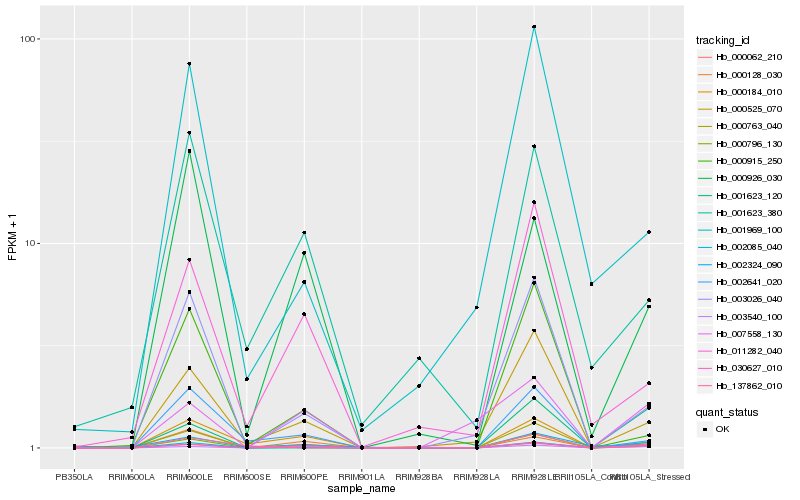
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_210 |
0.0 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Pyrus x bretschneideri] |
| 2 |
Hb_030627_010 |
0.1284295531 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |
| 3 |
Hb_000128_030 |
0.175317367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000796_130 |
0.1997742787 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 5 |
Hb_000525_070 |
0.2131706315 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like [Jatropha curcas] |
| 6 |
Hb_003026_040 |
0.2245924659 |
- |
- |
Uncharacterized protein TCM_007861 [Theobroma cacao] |
| 7 |
Hb_000763_040 |
0.2382286695 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 8 |
Hb_002085_040 |
0.2432710372 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 9 |
Hb_000184_010 |
0.2519898821 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 10 |
Hb_002641_020 |
0.2544498394 |
- |
- |
3'-5' exonuclease, putative [Ricinus communis] |
| 11 |
Hb_000926_030 |
0.2578330631 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_001623_120 |
0.2622680048 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X2 [Jatropha curcas] |
| 13 |
Hb_011282_040 |
0.2709429122 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 14 |
Hb_007558_130 |
0.2731714765 |
- |
- |
- |
| 15 |
Hb_002324_090 |
0.2790694985 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 16 |
Hb_001969_100 |
0.2806678915 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 17 |
Hb_000915_250 |
0.2812537823 |
- |
- |
chloroplast latex aldolase-like protein [Manihot esculenta] |
| 18 |
Hb_001623_380 |
0.2828416424 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003540_100 |
0.2831003998 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 20 |
Hb_137862_010 |
0.2863943049 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |