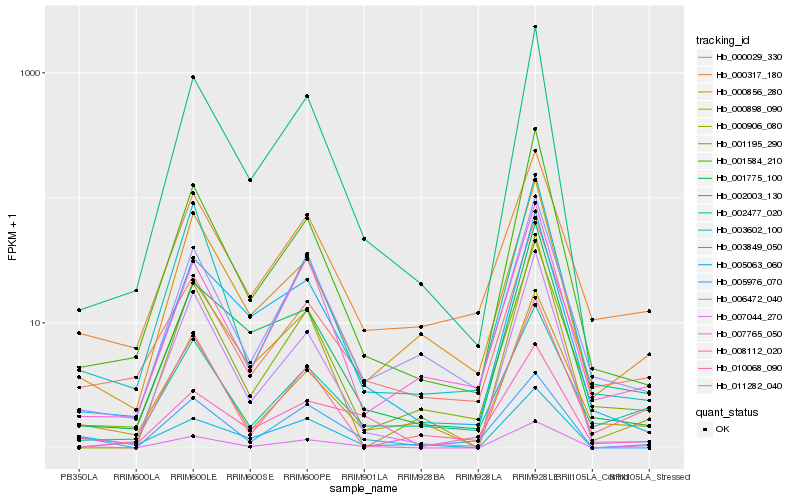
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007765_050 |
0.0 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001584_210 |
0.1761926446 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000898_090 |
0.1786332024 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105633600 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002003_130 |
0.1790056285 |
- |
- |
hypothetical protein CISIN_1g027843mg [Citrus sinensis] |
| 5 |
Hb_003849_050 |
0.1808150181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002477_020 |
0.1858358373 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |
| 7 |
Hb_001195_290 |
0.1859858519 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 8 |
Hb_000029_330 |
0.1866333768 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Jatropha curcas] |
| 9 |
Hb_007044_270 |
0.1874002232 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000856_280 |
0.191316979 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 11 |
Hb_010068_090 |
0.1919755106 |
- |
- |
hypothetical protein JCGZ_00885 [Jatropha curcas] |
| 12 |
Hb_003602_100 |
0.1932310607 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_005063_060 |
0.1938013337 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 14 |
Hb_008112_020 |
0.1967573105 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000906_080 |
0.1968770689 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_006472_040 |
0.1969639214 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001775_100 |
0.1977077443 |
- |
- |
PREDICTED: probable 2-carboxy-D-arabinitol-1-phosphatase [Jatropha curcas] |
| 18 |
Hb_000317_180 |
0.1979527446 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_011282_040 |
0.1987919322 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 20 |
Hb_005976_070 |
0.1990483305 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |