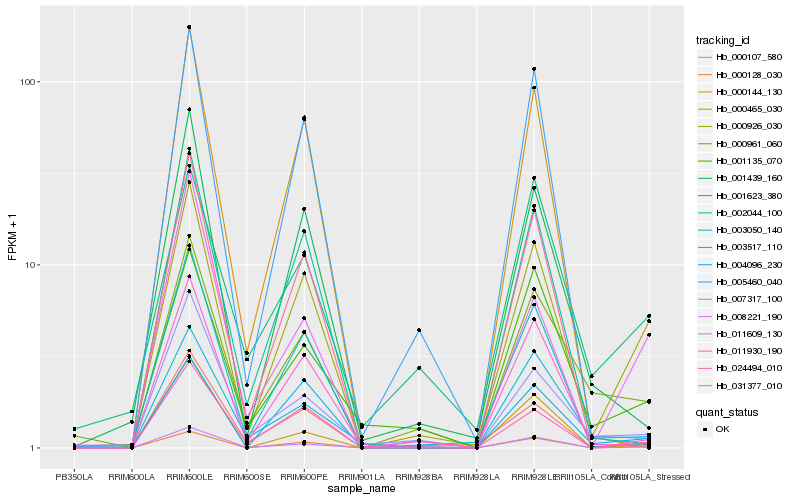
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000926_030 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_000128_030 |
0.1151174365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_024494_010 |
0.1553332134 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 4 |
Hb_000961_060 |
0.1693388074 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 5 |
Hb_004096_230 |
0.1720615072 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011609_130 |
0.1758585646 |
- |
- |
- |
| 7 |
Hb_008221_190 |
0.1788730072 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 8 |
Hb_001135_070 |
0.1831780566 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 9 |
Hb_003050_140 |
0.1859665118 |
- |
- |
PREDICTED: potassium channel KAT1 [Jatropha curcas] |
| 10 |
Hb_011930_190 |
0.1877876978 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003517_110 |
0.189580967 |
- |
- |
PREDICTED: uncharacterized protein LOC105633058 [Jatropha curcas] |
| 12 |
Hb_007317_100 |
0.1918469235 |
- |
- |
Peflin, putative [Ricinus communis] |
| 13 |
Hb_001439_160 |
0.192494435 |
- |
- |
PREDICTED: uncharacterized protein LOC105644097 [Jatropha curcas] |
| 14 |
Hb_000465_030 |
0.1952730285 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_000107_580 |
0.1979071452 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 16 |
Hb_031377_010 |
0.1979916167 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 17 |
Hb_001623_380 |
0.1980898372 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002044_100 |
0.2002536842 |
- |
- |
PREDICTED: uncharacterized protein LOC105643555 [Jatropha curcas] |
| 19 |
Hb_005460_040 |
0.2035704141 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 2 [Jatropha curcas] |
| 20 |
Hb_000144_130 |
0.2053310464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |