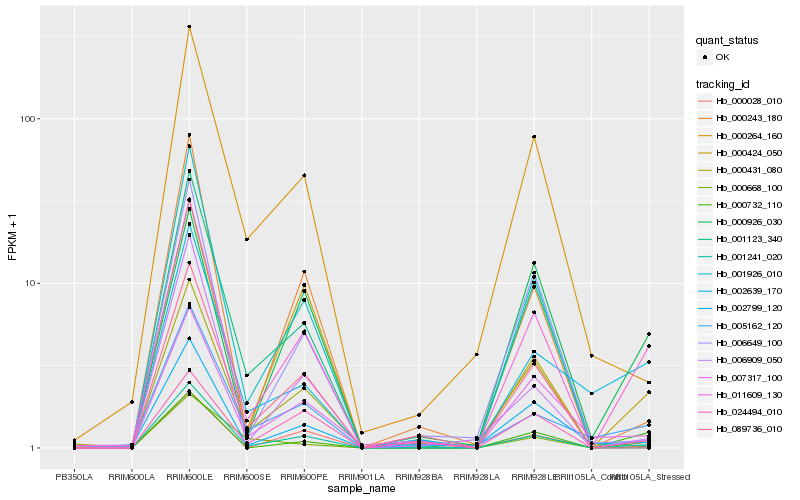
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011609_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_001241_020 |
0.1124895174 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 3 |
Hb_000732_110 |
0.1369991791 |
- |
- |
hypothetical protein JCGZ_21339 [Jatropha curcas] |
| 4 |
Hb_005162_120 |
0.1606222387 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 5 |
Hb_002639_170 |
0.1684288512 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 6 |
Hb_000028_010 |
0.1703544116 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_002799_120 |
0.171446236 |
- |
- |
PREDICTED: uncharacterized protein LOC105141197 [Populus euphratica] |
| 8 |
Hb_007317_100 |
0.1725078977 |
- |
- |
Peflin, putative [Ricinus communis] |
| 9 |
Hb_000926_030 |
0.1758585646 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000424_050 |
0.1780558001 |
- |
- |
erecta, putative [Ricinus communis] |
| 11 |
Hb_000243_180 |
0.1841872301 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1 [Jatropha curcas] |
| 12 |
Hb_024494_010 |
0.194921601 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 13 |
Hb_001926_010 |
0.1972268332 |
- |
- |
hypothetical protein Csa_2G383910 [Cucumis sativus] |
| 14 |
Hb_000264_160 |
0.1981134781 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 15 |
Hb_006649_100 |
0.1982479166 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 16 |
Hb_001123_340 |
0.1988733445 |
- |
- |
PREDICTED: uncharacterized protein LOC105123089 isoform X1 [Populus euphratica] |
| 17 |
Hb_000431_080 |
0.2000169926 |
- |
- |
PREDICTED: uncharacterized protein LOC105632547 [Jatropha curcas] |
| 18 |
Hb_000668_100 |
0.2001681337 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 19 |
Hb_089736_010 |
0.2009131157 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 7 [Populus euphratica] |
| 20 |
Hb_006909_050 |
0.2013959501 |
transcription factor |
TF Family: OFP |
hypothetical protein POPTR_0008s01840g [Populus trichocarpa] |