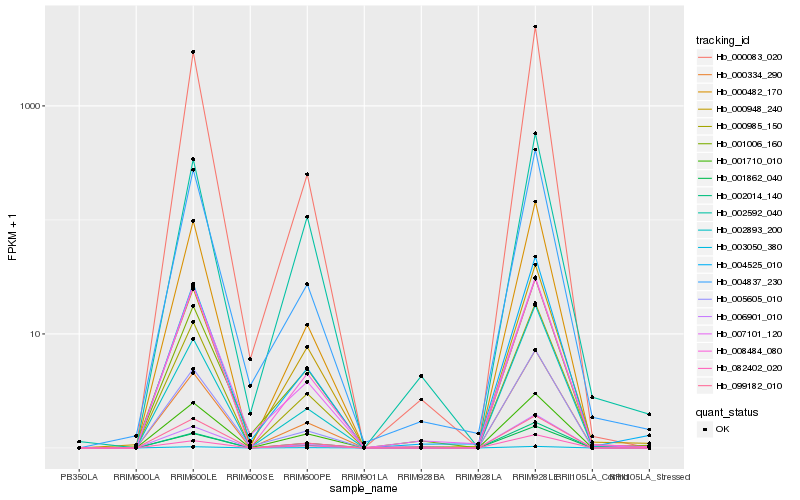
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000985_150 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 2 |
Hb_000334_290 |
0.0422546247 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 3 |
Hb_000482_170 |
0.0586379118 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 4 |
Hb_008484_080 |
0.0615433268 |
- |
- |
PREDICTED: crocetin glucosyltransferase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_004525_010 |
0.0615641531 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 6 |
Hb_000948_240 |
0.0653095436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006901_010 |
0.0667721767 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 8 |
Hb_004837_230 |
0.0677484123 |
- |
- |
unknown [Populus trichocarpa] |
| 9 |
Hb_002014_140 |
0.0718713404 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 10 |
Hb_005605_010 |
0.0745556214 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 11 |
Hb_003050_380 |
0.0747423072 |
- |
- |
hypothetical protein POPTR_0007s03380g [Populus trichocarpa] |
| 12 |
Hb_099182_010 |
0.0748735532 |
- |
- |
PREDICTED: uncharacterized protein LOC101214783 [Cucumis sativus] |
| 13 |
Hb_007101_120 |
0.0756884905 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 14 |
Hb_001862_040 |
0.0757747359 |
- |
- |
heat shock protein 70 (HSP70)-interacting protein, putative [Ricinus communis] |
| 15 |
Hb_002893_200 |
0.0777470376 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_082402_020 |
0.0778142369 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 17 |
Hb_001710_010 |
0.0778867025 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 18 |
Hb_002592_040 |
0.0780909357 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 19 |
Hb_001006_160 |
0.079219174 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_000083_020 |
0.0824060962 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |