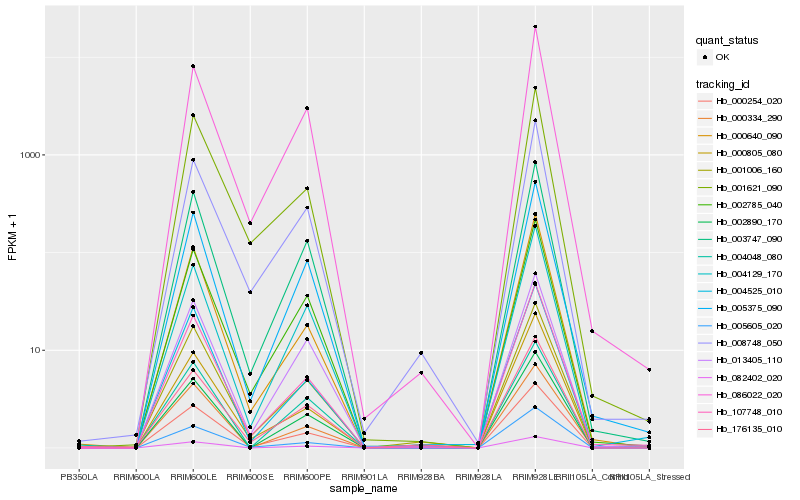
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000254_020 |
0.0 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 2 |
Hb_107748_010 |
0.0663818166 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_005375_090 |
0.0715518458 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003747_090 |
0.0715804268 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_001006_160 |
0.0735763626 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_002785_040 |
0.0738634084 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 7 |
Hb_004525_010 |
0.0749378205 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 8 |
Hb_005605_020 |
0.0781522848 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 9 |
Hb_086022_020 |
0.0807225943 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 10 |
Hb_004129_170 |
0.0837451685 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 11 |
Hb_002890_170 |
0.0838869109 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 12 |
Hb_176135_010 |
0.0844219859 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_082402_020 |
0.0845948696 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 14 |
Hb_000334_290 |
0.0847036372 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 15 |
Hb_000640_090 |
0.0847996477 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_001621_090 |
0.0851361296 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 17 |
Hb_008748_050 |
0.0851389937 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 18 |
Hb_000805_080 |
0.0851800512 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_013405_110 |
0.0861548552 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 20 |
Hb_004048_080 |
0.0863341098 |
- |
- |
hypothetical protein POPTR_0001s40360g [Populus trichocarpa] |