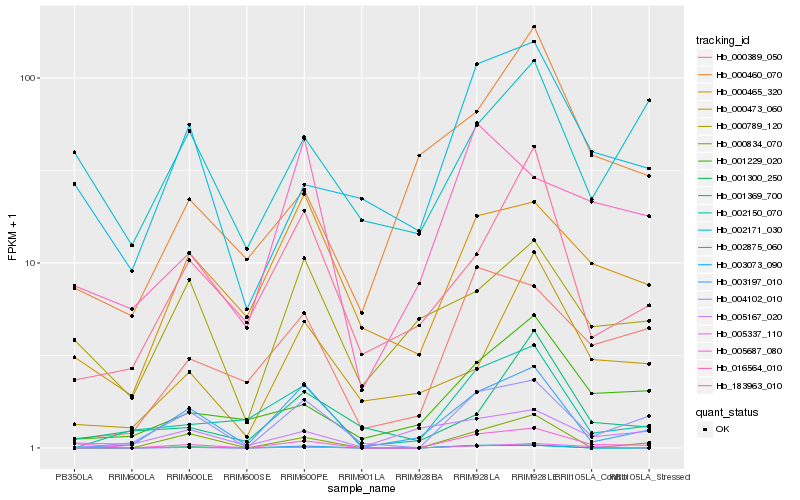
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005687_080 |
0.0 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 2 |
Hb_003073_090 |
0.2618019367 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_0004s23920g [Populus trichocarpa] |
| 3 |
Hb_002875_060 |
0.2671891663 |
- |
- |
hypothetical protein L484_023461 [Morus notabilis] |
| 4 |
Hb_004102_010 |
0.2840579152 |
- |
- |
F-box family protein [Theobroma cacao] |
| 5 |
Hb_005337_110 |
0.2874791957 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 6 |
Hb_003197_010 |
0.2989284549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001229_020 |
0.2989512518 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000834_070 |
0.3032180652 |
- |
- |
PREDICTED: tankyrase-1 isoform X4 [Jatropha curcas] |
| 9 |
Hb_000389_050 |
0.3097458006 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 10 |
Hb_002150_070 |
0.3164364636 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 11 |
Hb_000460_070 |
0.3251843218 |
- |
- |
luminal binding protein [Gossypium hirsutum] |
| 12 |
Hb_016564_010 |
0.3285499647 |
- |
- |
PREDICTED: uncharacterized protein LOC101764501 [Setaria italica] |
| 13 |
Hb_001300_250 |
0.3294759282 |
- |
- |
hypothetical protein JCGZ_22020 [Jatropha curcas] |
| 14 |
Hb_183963_010 |
0.3309676787 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |
| 15 |
Hb_000465_320 |
0.3320391046 |
- |
- |
PREDICTED: nudix hydrolase 8 [Jatropha curcas] |
| 16 |
Hb_000473_060 |
0.3328147302 |
- |
- |
PREDICTED: psbP domain-containing protein 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001369_700 |
0.3330655614 |
- |
- |
- |
| 18 |
Hb_005167_020 |
0.3351881519 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 19 |
Hb_000789_120 |
0.3361858136 |
- |
- |
PREDICTED: derlin-1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002171_030 |
0.3374901608 |
- |
- |
plastid acyl-ACP thioesterase [Vernicia fordii] |