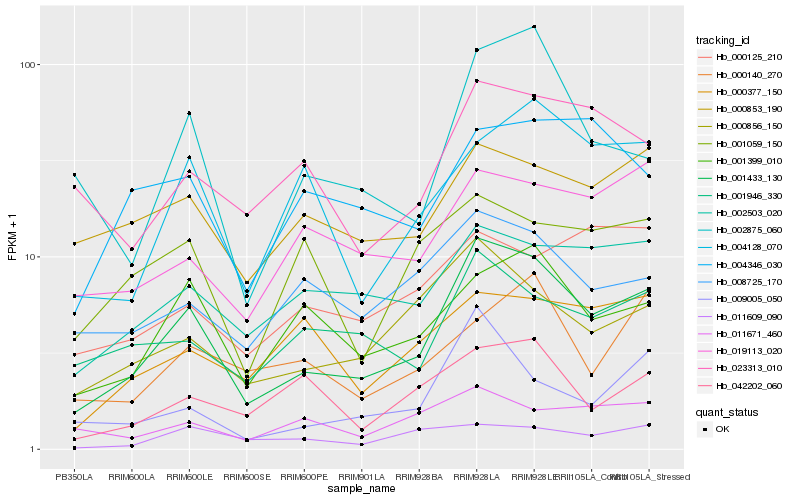
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001433_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647468 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000856_150 |
0.1629265614 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 3 |
Hb_001399_010 |
0.1756421669 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_002503_020 |
0.1783520587 |
- |
- |
Phosphatase 2C family protein, putative [Theobroma cacao] |
| 5 |
Hb_002875_060 |
0.1856059119 |
- |
- |
hypothetical protein L484_023461 [Morus notabilis] |
| 6 |
Hb_001946_330 |
0.1864232588 |
- |
- |
PREDICTED: transmembrane protein 128 [Jatropha curcas] |
| 7 |
Hb_000853_190 |
0.1882086523 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 8 |
Hb_023313_010 |
0.189095699 |
- |
- |
PREDICTED: linolenate hydroperoxide lyase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_004128_070 |
0.1891460481 |
- |
- |
PREDICTED: thioredoxin-like protein AAED1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_009005_050 |
0.189169697 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 11 |
Hb_008725_170 |
0.1903108676 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Populus euphratica] |
| 12 |
Hb_019113_020 |
0.193574578 |
- |
- |
hypothetical protein JCGZ_13398 [Jatropha curcas] |
| 13 |
Hb_042202_060 |
0.1985152558 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Jatropha curcas] |
| 14 |
Hb_001059_150 |
0.2024261896 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 15 |
Hb_004346_030 |
0.2025058866 |
- |
- |
sucrose phosphate phosphatase, putative [Ricinus communis] |
| 16 |
Hb_000377_150 |
0.2026949069 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 17 |
Hb_011671_460 |
0.2059561671 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 18 |
Hb_000125_210 |
0.2086603511 |
- |
- |
PREDICTED: electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_011609_090 |
0.2101373405 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_000140_270 |
0.2104402049 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |