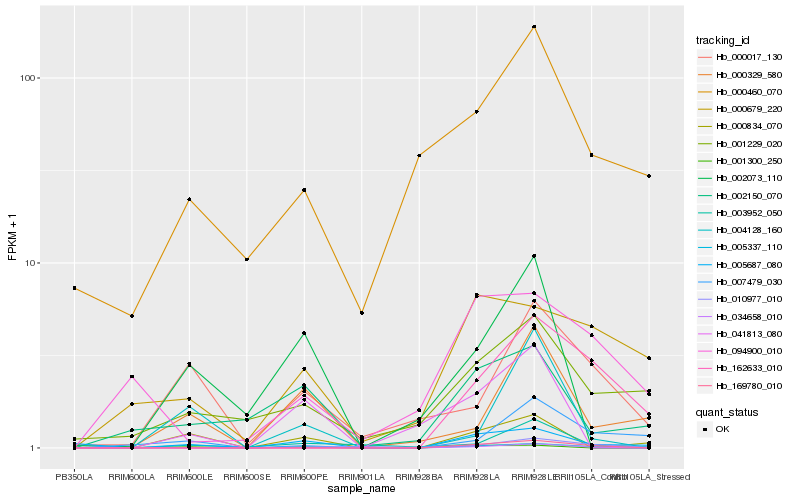
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005337_110 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_169780_010 |
0.2757008083 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 3 |
Hb_034658_010 |
0.2872727957 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_005687_080 |
0.2874791957 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 5 |
Hb_000017_130 |
0.3206425981 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2-like [Jatropha curcas] |
| 6 |
Hb_001300_250 |
0.3300938793 |
- |
- |
hypothetical protein JCGZ_22020 [Jatropha curcas] |
| 7 |
Hb_003952_050 |
0.3342834818 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 8 |
Hb_001229_020 |
0.3382251503 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_041813_080 |
0.340546159 |
- |
- |
- |
| 10 |
Hb_162633_010 |
0.3422428577 |
- |
- |
hypothetical protein POPTR_0011s02600g [Populus trichocarpa] |
| 11 |
Hb_010977_010 |
0.3440536952 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_000460_070 |
0.3460052025 |
- |
- |
luminal binding protein [Gossypium hirsutum] |
| 13 |
Hb_000679_220 |
0.3461354819 |
- |
- |
PREDICTED: uncharacterized protein LOC105637638 [Jatropha curcas] |
| 14 |
Hb_007479_030 |
0.3488172896 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 15 |
Hb_004128_160 |
0.3494273593 |
- |
- |
PREDICTED: uncharacterized protein LOC105639161 [Jatropha curcas] |
| 16 |
Hb_002073_110 |
0.3513261594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002150_070 |
0.3517154446 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 18 |
Hb_000329_580 |
0.3520360989 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_094900_010 |
0.3550073445 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000834_070 |
0.3582840763 |
- |
- |
PREDICTED: tankyrase-1 isoform X4 [Jatropha curcas] |