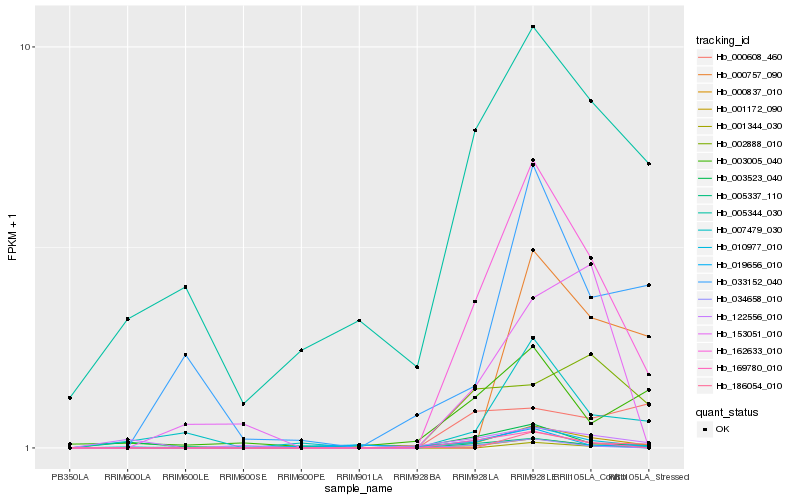
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162633_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s02600g [Populus trichocarpa] |
| 2 |
Hb_169780_010 |
0.2125975568 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 3 |
Hb_034658_010 |
0.2293400162 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_001172_090 |
0.2402502077 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 5 |
Hb_002888_010 |
0.2596304191 |
- |
- |
- |
| 6 |
Hb_007479_030 |
0.2898533268 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 7 |
Hb_010977_010 |
0.2967125358 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_000608_460 |
0.3130014949 |
- |
- |
- |
| 9 |
Hb_001344_030 |
0.3197316815 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_019656_010 |
0.3214470831 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_000837_010 |
0.3258379985 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_003005_040 |
0.3275971452 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_033152_040 |
0.3325529581 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 14 |
Hb_003523_040 |
0.3358462858 |
- |
- |
F15O4.13 [Arabidopsis thaliana] |
| 15 |
Hb_000757_090 |
0.3369399085 |
- |
- |
SPla/RYanodine receptor domain-containing protein isoform 3 [Theobroma cacao] |
| 16 |
Hb_153051_010 |
0.3375940503 |
- |
- |
- |
| 17 |
Hb_186054_010 |
0.339197754 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_005344_030 |
0.3414437139 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Jatropha curcas] |
| 19 |
Hb_005337_110 |
0.3422428577 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 20 |
Hb_122556_010 |
0.3471231709 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |