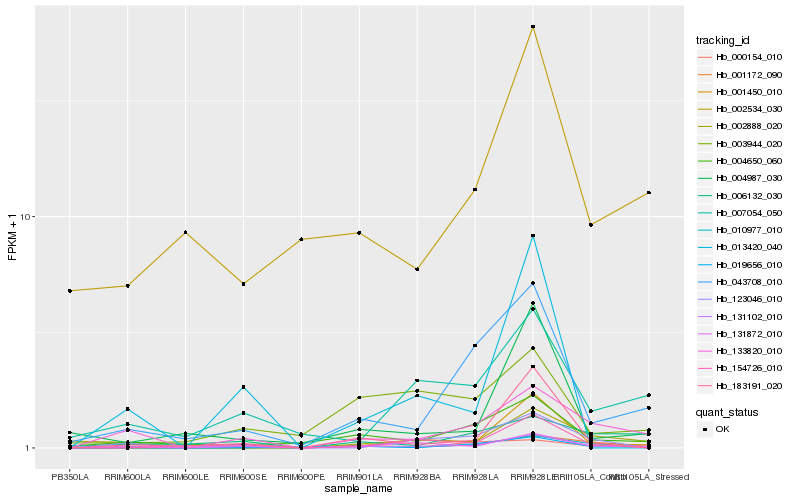
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_131102_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_133820_010 |
0.1724295362 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 3 |
Hb_131872_010 |
0.2024094154 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_019656_010 |
0.210044353 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_043708_010 |
0.2120793664 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 6 |
Hb_001450_010 |
0.2279788774 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 7 |
Hb_004650_060 |
0.2355566744 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 8 |
Hb_007054_050 |
0.2500817923 |
- |
- |
- |
| 9 |
Hb_183191_020 |
0.2602558977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_123046_010 |
0.2645669073 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 11 |
Hb_000154_010 |
0.2694398466 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_010977_010 |
0.2729223803 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_002534_030 |
0.2803123311 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 14 |
Hb_002888_020 |
0.2806391035 |
- |
- |
- |
| 15 |
Hb_006132_030 |
0.282114833 |
- |
- |
hypothetical protein CARUB_v10012126mg, partial [Capsella rubella] |
| 16 |
Hb_001172_090 |
0.2832953898 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 17 |
Hb_154726_010 |
0.2869577423 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 18 |
Hb_013420_040 |
0.2878032219 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_004987_030 |
0.2883019917 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 20 |
Hb_003944_020 |
0.2902656448 |
- |
- |
PREDICTED: uncharacterized protein LOC105110580 [Populus euphratica] |