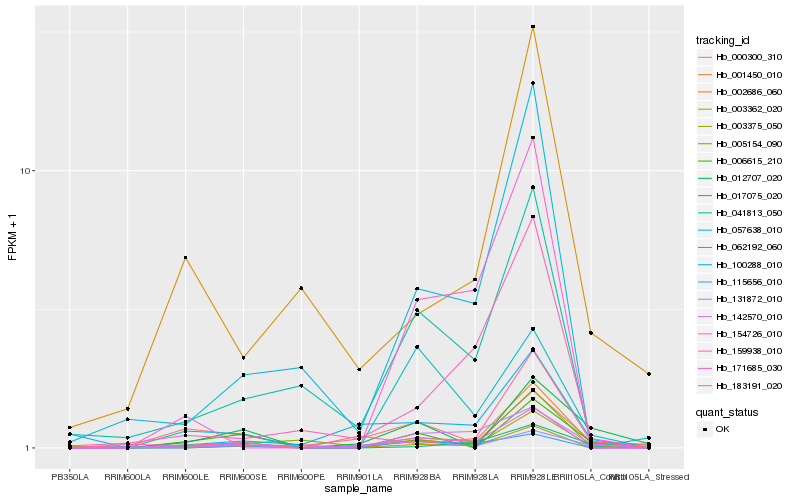
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_154726_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 2 |
Hb_001450_010 |
0.22345458 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 3 |
Hb_003375_050 |
0.232233366 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_041813_050 |
0.2426102096 |
- |
- |
hypothetical protein NitaMp096 [Nicotiana tabacum] |
| 5 |
Hb_017075_020 |
0.2426795048 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 6 |
Hb_057638_010 |
0.2435610033 |
- |
- |
PREDICTED: uncharacterized protein LOC105637008 [Jatropha curcas] |
| 7 |
Hb_100288_010 |
0.2545599299 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_183191_020 |
0.260763477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_171685_030 |
0.2624404914 |
- |
- |
URF-1 [Citrullus lanatus] |
| 10 |
Hb_062192_060 |
0.2633384387 |
- |
- |
maturase (mitochondrion) [Hevea brasiliensis] |
| 11 |
Hb_003362_020 |
0.2664842296 |
- |
- |
PREDICTED: uncharacterized protein LOC104098435, partial [Nicotiana tomentosiformis] |
| 12 |
Hb_115656_010 |
0.2665951773 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 13 |
Hb_006615_210 |
0.2666597828 |
- |
- |
PREDICTED: uncharacterized protein LOC105640677 isoform X2 [Jatropha curcas] |
| 14 |
Hb_131872_010 |
0.2686642108 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_000300_310 |
0.2698798397 |
- |
- |
PREDICTED: uncharacterized protein LOC104896902 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_012707_020 |
0.2710006048 |
- |
- |
- |
| 17 |
Hb_002686_060 |
0.2717722215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_142570_010 |
0.2718440532 |
- |
- |
PREDICTED: uncharacterized protein LOC104907573 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_159938_010 |
0.2719090074 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 20 |
Hb_005154_090 |
0.2720779737 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |