| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
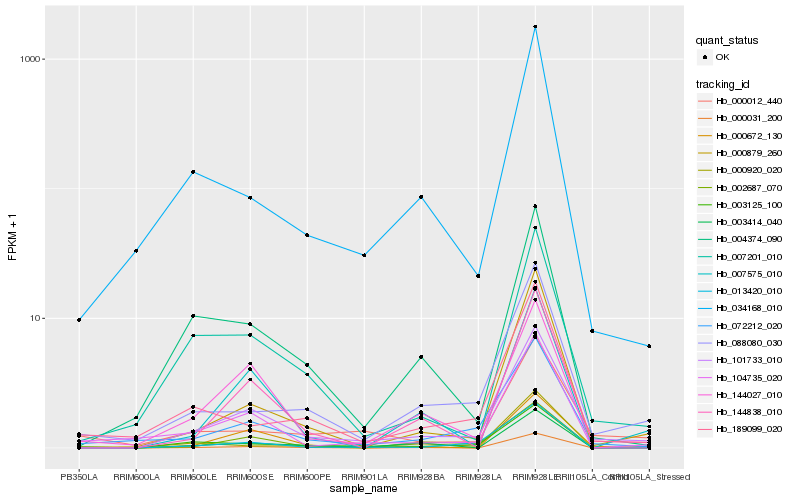
Hb_101733_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_072212_020 |
0.1586367352 |
- |
- |
hypothetical protein B456_001G162300 [Gossypium raimondii] |
| 3 |
Hb_144838_010 |
0.1688709945 |
- |
- |
hypothetical protein JCGZ_08246 [Jatropha curcas] |
| 4 |
Hb_088080_030 |
0.1689047926 |
- |
- |
ribosomal protein S4 [Hevea brasiliensis] |
| 5 |
Hb_000879_260 |
0.1714041863 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 6 |
Hb_007201_010 |
0.1732573068 |
- |
- |
PREDICTED: uncharacterized protein LOC105631226 isoform X2 [Jatropha curcas] |
| 7 |
Hb_013420_010 |
0.1800861394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_034168_010 |
0.1809227839 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 9 |
Hb_004374_090 |
0.1847331931 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 10 |
Hb_003414_040 |
0.1856373688 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 11 |
Hb_104735_020 |
0.1867148159 |
- |
- |
ATPase subunit 4 (mitochondrion) [Hevea brasiliensis] |
| 12 |
Hb_000672_130 |
0.1885180802 |
- |
- |
hypothetical protein POPTR_0010s22870g [Populus trichocarpa] |
| 13 |
Hb_000012_440 |
0.189924218 |
- |
- |
PREDICTED: uncharacterized protein LOC104248335 [Nicotiana sylvestris] |
| 14 |
Hb_003125_100 |
0.1909303788 |
- |
- |
NAD(P)-binding Rossmann-fold superfamily protein [Theobroma cacao] |
| 15 |
Hb_007575_010 |
0.1922033935 |
- |
- |
PREDICTED: cytochrome P450 71A1 [Vitis vinifera] |
| 16 |
Hb_000031_200 |
0.1944367417 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM3-like [Jatropha curcas] |
| 17 |
Hb_000920_020 |
0.1947784074 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 18 |
Hb_189099_020 |
0.1958736899 |
- |
- |
NADH dehydrogenase subunit 9 (mitochondrion) [Hevea brasiliensis] |
| 19 |
Hb_002687_070 |
0.1961800755 |
- |
- |
polyprotein [Ananas comosus] |
| 20 |
Hb_144027_010 |
0.1972668514 |
- |
- |
- |