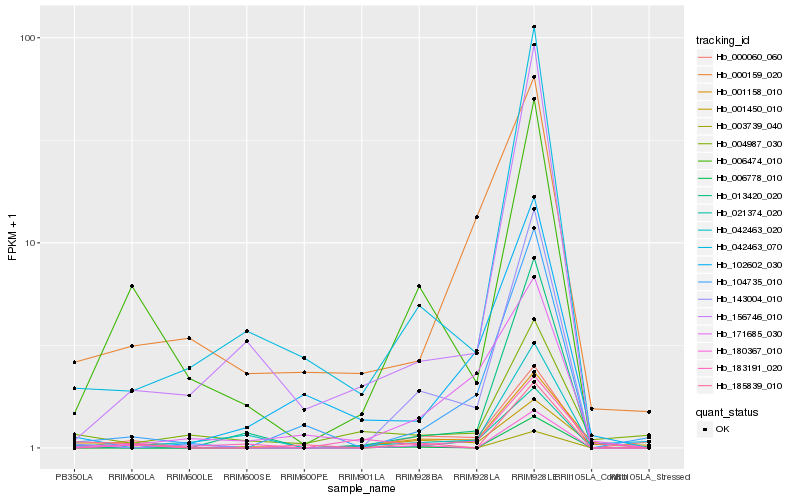
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001158_010 |
0.0 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 2 |
Hb_006474_010 |
0.2375260761 |
- |
- |
hypothetical protein VITISV_032357 [Vitis vinifera] |
| 3 |
Hb_183191_020 |
0.2376850143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_143004_010 |
0.24179404 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C1071.09c [Eucalyptus grandis] |
| 5 |
Hb_001450_010 |
0.2439334915 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 6 |
Hb_000060_060 |
0.2491061691 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_22940 [Jatropha curcas] |
| 7 |
Hb_004987_030 |
0.2531694548 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 8 |
Hb_000159_020 |
0.2557333675 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Jatropha curcas] |
| 9 |
Hb_013420_020 |
0.2611821267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_104735_010 |
0.2616339066 |
- |
- |
cytochrome c biogenesis C (mitochondrion) [Hevea brasiliensis] |
| 11 |
Hb_042463_070 |
0.2629315058 |
- |
- |
60S ribosomal protein L16, putative [Ricinus communis] |
| 12 |
Hb_171685_030 |
0.2630111146 |
- |
- |
URF-1 [Citrullus lanatus] |
| 13 |
Hb_185839_010 |
0.2650822316 |
- |
- |
- |
| 14 |
Hb_021374_020 |
0.2698405043 |
- |
- |
PREDICTED: uncharacterized protein LOC101298847, partial [Fragaria vesca subsp. vesca] |
| 15 |
Hb_003739_040 |
0.2702919684 |
- |
- |
RNA-directed DNA polymerase (Reverse transcriptase); Chromo; Zinc finger, CCHC-type; Peptidase aspartic, active site; Polynucleotidyl transferase, Ribonuclease H fold [Medicago truncatula] |
| 16 |
Hb_102602_030 |
0.2711824638 |
- |
- |
hypothetical protein (mitochondrion) [Vicia faba] |
| 17 |
Hb_006778_010 |
0.2751694635 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 18 |
Hb_156746_010 |
0.2755848155 |
- |
- |
ATP synthase F1 subunit 1 (mitochondrion) [Hevea brasiliensis] |
| 19 |
Hb_180367_010 |
0.2763416842 |
- |
- |
PREDICTED: uncharacterized protein LOC105794100 [Gossypium raimondii] |
| 20 |
Hb_042463_020 |
0.2772920794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |