| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
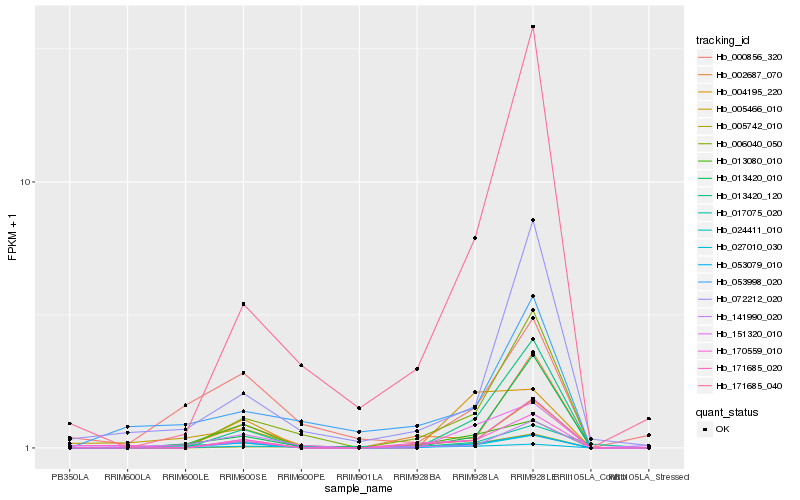
Hb_053079_010 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_013080_010 |
0.2186455335 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_024411_010 |
0.2192135408 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_006040_050 |
0.231917822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005466_010 |
0.244624233 |
- |
- |
ATPase subunit 6, partial (mitochondrion) [Celastrus scandens] |
| 6 |
Hb_027010_030 |
0.2492898301 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 7 |
Hb_151320_010 |
0.2540061187 |
- |
- |
- |
| 8 |
Hb_141990_020 |
0.2561882986 |
- |
- |
RecName: Full=Genome polyprotein; Contains: RecName: Full=Movement protein; Short=MP; Contains: RecName: Full=Capsid protein; Short=CP; Contains: RecName: Full=Aspartic protease; Short=PR; Contains: RecName: Full=Reverse transcriptase; Short=RT [Petunia vein clearing virus isolate Hohn] |
| 9 |
Hb_000856_320 |
0.2572156882 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |
| 10 |
Hb_002687_070 |
0.2581948539 |
- |
- |
polyprotein [Ananas comosus] |
| 11 |
Hb_053998_020 |
0.2606813486 |
- |
- |
- |
| 12 |
Hb_171685_020 |
0.260724853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_013420_120 |
0.2646612961 |
- |
- |
- |
| 14 |
Hb_072212_020 |
0.2713569054 |
- |
- |
hypothetical protein B456_001G162300 [Gossypium raimondii] |
| 15 |
Hb_005742_010 |
0.27260661 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 16 |
Hb_013420_010 |
0.2748303631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004195_220 |
0.2808883225 |
- |
- |
hypothetical protein PRUPE_ppa005288mg [Prunus persica] |
| 18 |
Hb_170559_010 |
0.283059843 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 19 |
Hb_171685_040 |
0.2855302544 |
- |
- |
ribosomal protein S13 (mitochondrion) [Hevea brasiliensis] |
| 20 |
Hb_017075_020 |
0.2870840151 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |