| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
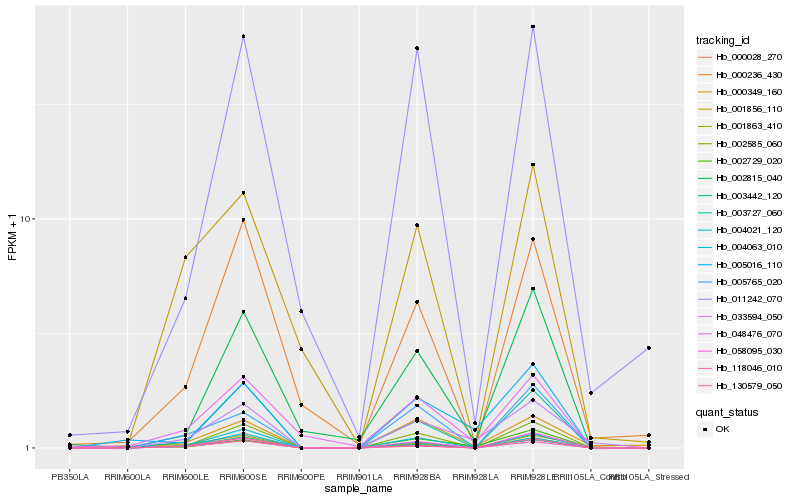
Hb_033594_050 |
0.0 |
- |
- |
PREDICTED: cyclin-D4-1-like [Jatropha curcas] |
| 2 |
Hb_000349_160 |
0.1719881023 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Citrus sinensis] |
| 3 |
Hb_005016_110 |
0.2002344266 |
- |
- |
- |
| 4 |
Hb_002585_060 |
0.2106311793 |
- |
- |
PREDICTED: uncharacterized protein LOC104899128, partial [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_004063_010 |
0.2110894555 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000028_270 |
0.2127718897 |
- |
- |
- |
| 7 |
Hb_058095_030 |
0.2133965581 |
- |
- |
- |
| 8 |
Hb_118046_010 |
0.216512821 |
- |
- |
PREDICTED: uncharacterized protein LOC102596706 [Solanum tuberosum] |
| 9 |
Hb_003727_060 |
0.2219081783 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_048476_070 |
0.2240228986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003442_120 |
0.2247772971 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |
| 12 |
Hb_002815_040 |
0.2356700034 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 13 |
Hb_011242_070 |
0.2360294114 |
- |
- |
beta-cyanoalanine synthase [Hevea brasiliensis] |
| 14 |
Hb_000236_430 |
0.2370837517 |
- |
- |
PREDICTED: uncharacterized protein LOC105633302 [Jatropha curcas] |
| 15 |
Hb_005765_020 |
0.2395465672 |
- |
- |
- |
| 16 |
Hb_002729_020 |
0.2432243102 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |
| 17 |
Hb_004021_120 |
0.2450164642 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 18 |
Hb_001863_410 |
0.2466440686 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 19 |
Hb_130579_050 |
0.2548529883 |
- |
- |
hypothetical protein JCGZ_25577 [Jatropha curcas] |
| 20 |
Hb_001856_110 |
0.2562126069 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |