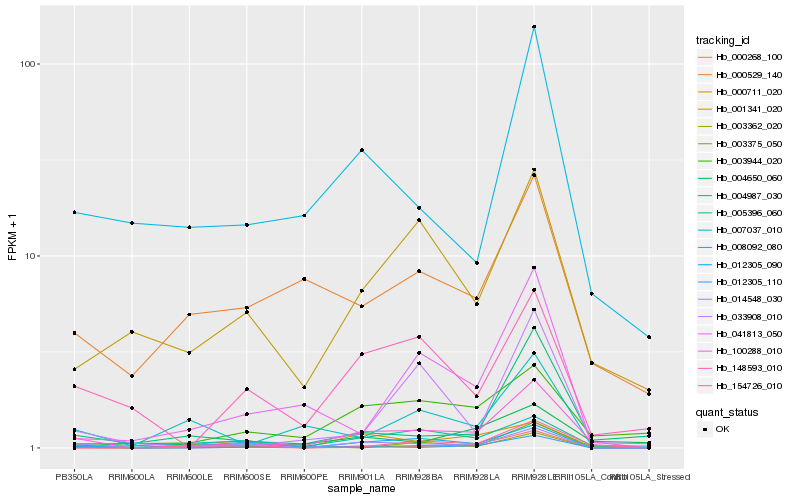
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_100288_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_014548_030 |
0.2142420473 |
- |
- |
PREDICTED: uncharacterized protein LOC104897580 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_005396_060 |
0.2428391208 |
- |
- |
- |
| 4 |
Hb_012305_110 |
0.2437687638 |
- |
- |
PREDICTED: uncharacterized protein LOC105156417 [Sesamum indicum] |
| 5 |
Hb_012305_090 |
0.2507457131 |
- |
- |
hypothetical protein VITISV_018166 [Vitis vinifera] |
| 6 |
Hb_003362_020 |
0.254060275 |
- |
- |
PREDICTED: uncharacterized protein LOC104098435, partial [Nicotiana tomentosiformis] |
| 7 |
Hb_004987_030 |
0.2544551585 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 8 |
Hb_154726_010 |
0.2545599299 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 9 |
Hb_008092_080 |
0.2612378061 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 10 |
Hb_003944_020 |
0.261688461 |
- |
- |
PREDICTED: uncharacterized protein LOC105110580 [Populus euphratica] |
| 11 |
Hb_041813_050 |
0.2624203507 |
- |
- |
hypothetical protein NitaMp096 [Nicotiana tabacum] |
| 12 |
Hb_001341_020 |
0.2626040105 |
- |
- |
Uncharacterised protein [Bordetella pertussis] |
| 13 |
Hb_000268_100 |
0.2657966334 |
- |
- |
- |
| 14 |
Hb_148593_010 |
0.2754176528 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR-like [Jatropha curcas] |
| 15 |
Hb_033908_010 |
0.2776751468 |
- |
- |
- |
| 16 |
Hb_007037_010 |
0.2786833322 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 17 |
Hb_003375_050 |
0.2800183521 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_000711_020 |
0.2811540875 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 19 |
Hb_004650_060 |
0.2843266938 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 20 |
Hb_000529_140 |
0.285100525 |
- |
- |
conserved hypothetical protein [Ricinus communis] |