| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
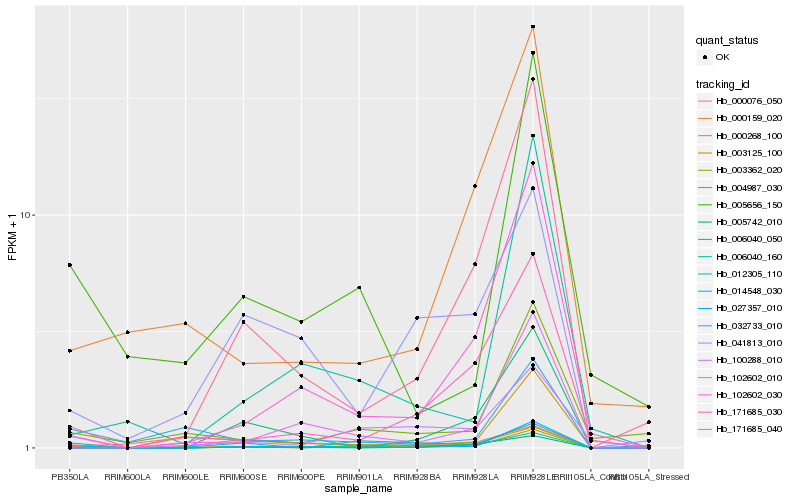
Hb_012305_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105156417 [Sesamum indicum] |
| 2 |
Hb_102602_030 |
0.2400990568 |
- |
- |
hypothetical protein (mitochondrion) [Vicia faba] |
| 3 |
Hb_100288_010 |
0.2437687638 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_000076_050 |
0.2579372044 |
- |
- |
PREDICTED: uncharacterized protein LOC105155919 [Sesamum indicum] |
| 5 |
Hb_171685_040 |
0.2600741987 |
- |
- |
ribosomal protein S13 (mitochondrion) [Hevea brasiliensis] |
| 6 |
Hb_005656_150 |
0.2712810932 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 7 |
Hb_102602_010 |
0.2713834252 |
- |
- |
orf175 (mitochondrion) [Panax ginseng] |
| 8 |
Hb_000159_020 |
0.2759532392 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Jatropha curcas] |
| 9 |
Hb_003362_020 |
0.2788969151 |
- |
- |
PREDICTED: uncharacterized protein LOC104098435, partial [Nicotiana tomentosiformis] |
| 10 |
Hb_014548_030 |
0.2805320246 |
- |
- |
PREDICTED: uncharacterized protein LOC104897580 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_000268_100 |
0.2806283925 |
- |
- |
- |
| 12 |
Hb_032733_010 |
0.2960036296 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 13 |
Hb_004987_030 |
0.2967582309 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 14 |
Hb_171685_030 |
0.2976257811 |
- |
- |
URF-1 [Citrullus lanatus] |
| 15 |
Hb_041813_010 |
0.2993320905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005742_010 |
0.30015021 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 17 |
Hb_003125_100 |
0.3012295934 |
- |
- |
NAD(P)-binding Rossmann-fold superfamily protein [Theobroma cacao] |
| 18 |
Hb_006040_050 |
0.3037907447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006040_160 |
0.3039160911 |
- |
- |
ribosomal protein S12 (mitochondrion) [Hevea brasiliensis] |
| 20 |
Hb_027357_010 |
0.3056337815 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |