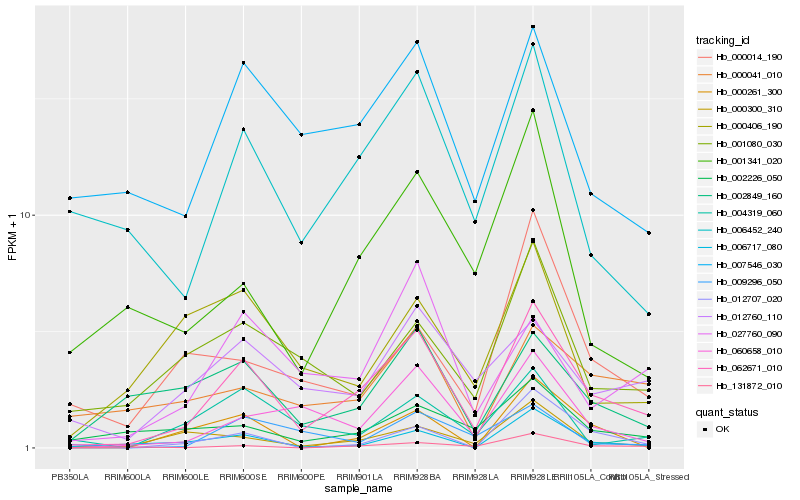
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_062671_010 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_060658_010 |
0.2200932958 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 3 |
Hb_131872_010 |
0.2238299142 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_006452_240 |
0.2263379116 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 15a isoform X1 [Jatropha curcas] |
| 5 |
Hb_027760_090 |
0.230537712 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000300_310 |
0.2316103428 |
- |
- |
PREDICTED: uncharacterized protein LOC104896902 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_012760_110 |
0.2318887108 |
- |
- |
PREDICTED: methyltransferase-like protein 23 isoform X1 [Jatropha curcas] |
| 8 |
Hb_006717_080 |
0.2332121038 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 9 |
Hb_002849_160 |
0.2338883813 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 10 |
Hb_007546_030 |
0.2365860366 |
- |
- |
transcription cofactor, putative [Ricinus communis] |
| 11 |
Hb_002226_050 |
0.2420490573 |
- |
- |
PREDICTED: uncharacterized protein LOC105641366 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001080_030 |
0.2445651235 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 13 |
Hb_001341_020 |
0.2450166863 |
- |
- |
Uncharacterised protein [Bordetella pertussis] |
| 14 |
Hb_000041_010 |
0.2514235908 |
- |
- |
pfkB-type carbohydrate kinase family protein [Populus trichocarpa] |
| 15 |
Hb_012707_020 |
0.2534151638 |
- |
- |
- |
| 16 |
Hb_004319_060 |
0.2539838623 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 17 |
Hb_000014_190 |
0.2564531965 |
- |
- |
Phytoene dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_009296_050 |
0.2569636258 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 19 |
Hb_000261_300 |
0.2584160661 |
- |
- |
- |
| 20 |
Hb_000406_190 |
0.2609417494 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |