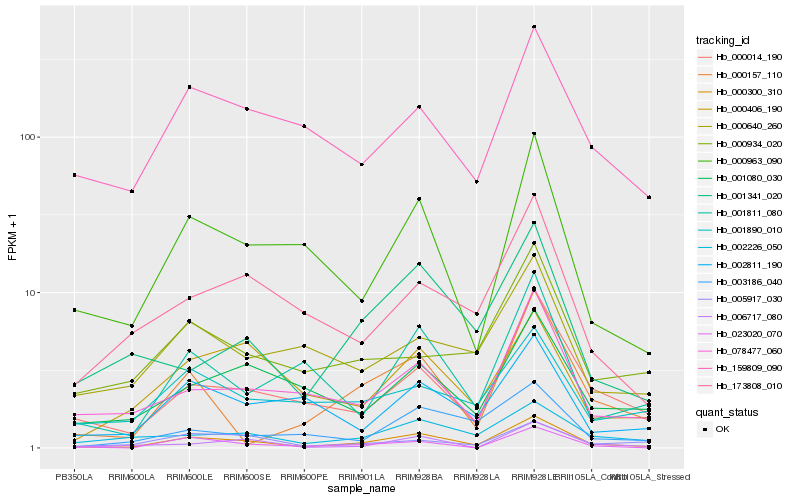
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_310 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104896902 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_000014_190 |
0.1665796242 |
- |
- |
Phytoene dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000963_090 |
0.1788649955 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_078477_060 |
0.1868494332 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 5 |
Hb_002226_050 |
0.200832987 |
- |
- |
PREDICTED: uncharacterized protein LOC105641366 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001811_080 |
0.2017797432 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 7 |
Hb_000157_110 |
0.2018770255 |
- |
- |
hypothetical protein Csa_6G439410 [Cucumis sativus] |
| 8 |
Hb_002811_190 |
0.203458419 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 9 |
Hb_023020_070 |
0.204396388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000406_190 |
0.204657936 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 11 |
Hb_173808_010 |
0.2058993478 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001080_030 |
0.2062069799 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 13 |
Hb_159809_090 |
0.2092498476 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 14 |
Hb_005917_030 |
0.210162086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006717_080 |
0.2105704807 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 16 |
Hb_001341_020 |
0.2123477678 |
- |
- |
Uncharacterised protein [Bordetella pertussis] |
| 17 |
Hb_001890_010 |
0.2143370059 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 18 |
Hb_003186_040 |
0.2152366645 |
- |
- |
PREDICTED: protein DENND6A [Jatropha curcas] |
| 19 |
Hb_000934_020 |
0.2188105149 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 20 |
Hb_000640_260 |
0.2192476796 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |