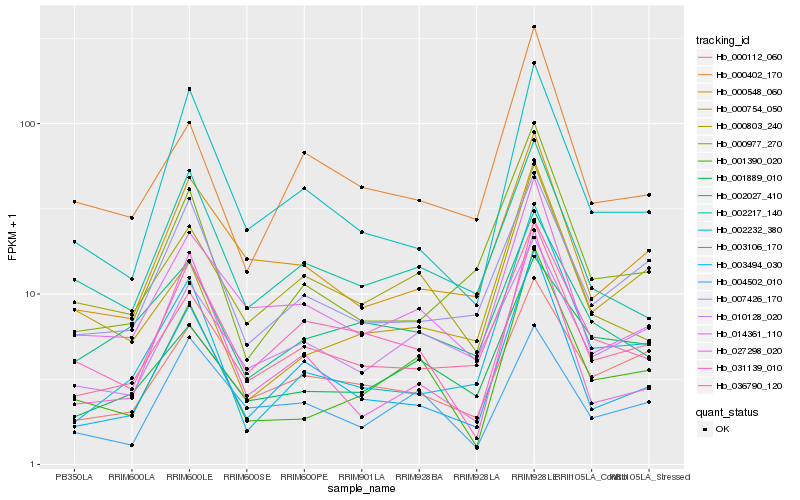
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001390_020 |
0.0 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_031139_010 |
0.162572476 |
- |
- |
PREDICTED: uncharacterized protein LOC105632403 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000754_050 |
0.165746268 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_014361_110 |
0.1683460486 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_002232_380 |
0.1706109399 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_003494_030 |
0.1708823902 |
- |
- |
PREDICTED: protease Do-like 8, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003106_170 |
0.171142297 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 8 |
Hb_036790_120 |
0.1740218379 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001889_010 |
0.1748389533 |
transcription factor |
TF Family: Orphans |
PREDICTED: protein CHLOROPLAST IMPORT APPARATUS 2 [Jatropha curcas] |
| 10 |
Hb_000803_240 |
0.1779758328 |
- |
- |
hypothetical protein CICLE_v100143612mg, partial [Citrus clementina] |
| 11 |
Hb_002217_140 |
0.1809412611 |
- |
- |
PREDICTED: 30S ribosomal protein S1, chloroplastic [Nelumbo nucifera] |
| 12 |
Hb_004502_010 |
0.182290811 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 13 |
Hb_000548_060 |
0.1827890786 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 14 |
Hb_000112_060 |
0.1854114975 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 15 |
Hb_007426_170 |
0.1856877238 |
transcription factor |
TF Family: TCP |
- |
| 16 |
Hb_000977_270 |
0.1861182239 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002027_410 |
0.1866200055 |
- |
- |
PREDICTED: protease Do-like 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_010128_020 |
0.1869954942 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 19 |
Hb_000402_170 |
0.1877616745 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 20 |
Hb_027298_020 |
0.1938428627 |
- |
- |
hypothetical protein POPTR_0005s05760g [Populus trichocarpa] |