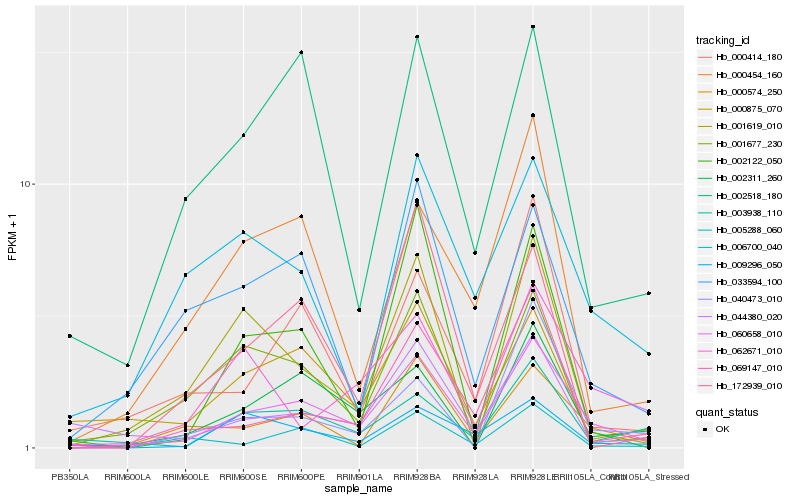
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_060658_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_000574_250 |
0.1812217448 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 3 |
Hb_002311_260 |
0.1912181432 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 4 |
Hb_000414_180 |
0.203483083 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 5 |
Hb_000454_160 |
0.2082609272 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_003938_110 |
0.2094953559 |
- |
- |
polyprotein [Ananas comosus] |
| 7 |
Hb_069147_010 |
0.2099087801 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At4g27220 isoform X1 [Populus euphratica] |
| 8 |
Hb_000875_070 |
0.2119901368 |
- |
- |
- |
| 9 |
Hb_033594_100 |
0.2139158565 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_001677_230 |
0.2141086383 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002518_180 |
0.2150087511 |
- |
- |
hypothetical protein POPTR_0001s25460g [Populus trichocarpa] |
| 12 |
Hb_044380_020 |
0.2153530132 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 13 |
Hb_005288_060 |
0.2195400448 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 14 |
Hb_009296_050 |
0.22008041 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 15 |
Hb_062671_010 |
0.2200932958 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001619_010 |
0.2249820257 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 17 |
Hb_002122_050 |
0.2271771396 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 18 |
Hb_040473_010 |
0.2273202839 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 19 |
Hb_172939_010 |
0.2285967872 |
- |
- |
unknown [Picea sitchensis] |
| 20 |
Hb_006700_040 |
0.2286956008 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |