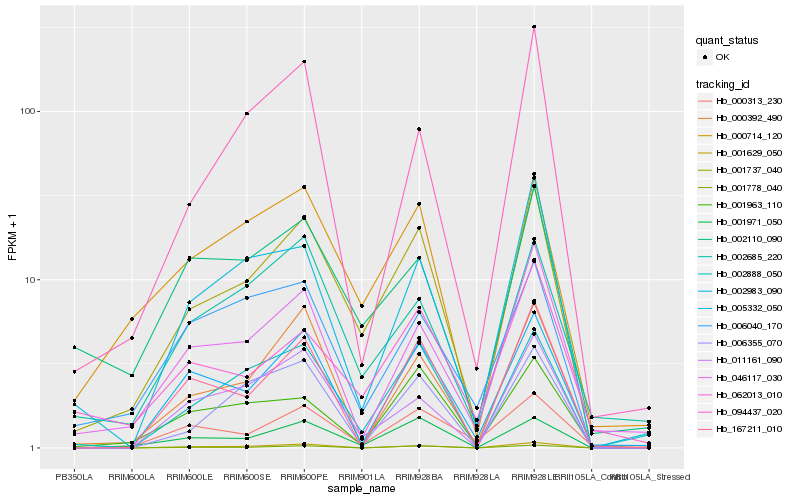
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001737_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 2 |
Hb_001971_050 |
0.1325971158 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 3 |
Hb_000714_120 |
0.1452251424 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 4 |
Hb_002888_050 |
0.150986389 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000392_490 |
0.1611959004 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005332_050 |
0.1693440419 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 7 |
Hb_002110_090 |
0.1720644163 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 8 |
Hb_001963_110 |
0.1720900581 |
- |
- |
- |
| 9 |
Hb_002685_220 |
0.1726275911 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_011161_090 |
0.172768378 |
- |
- |
- |
| 11 |
Hb_062013_010 |
0.1736799467 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 12 |
Hb_001778_040 |
0.1736813102 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |
| 13 |
Hb_046117_030 |
0.1738636206 |
- |
- |
sialyltransferase, putative [Ricinus communis] |
| 14 |
Hb_002983_090 |
0.176018746 |
- |
- |
Altered inheritance of mitochondria protein 32 [Glycine soja] |
| 15 |
Hb_000313_230 |
0.1767609324 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 16 |
Hb_001629_050 |
0.1774150845 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 17 |
Hb_167211_010 |
0.1774968097 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_006040_170 |
0.1793956859 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 19 |
Hb_006355_070 |
0.1802845574 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 20 |
Hb_094437_020 |
0.1809206514 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |