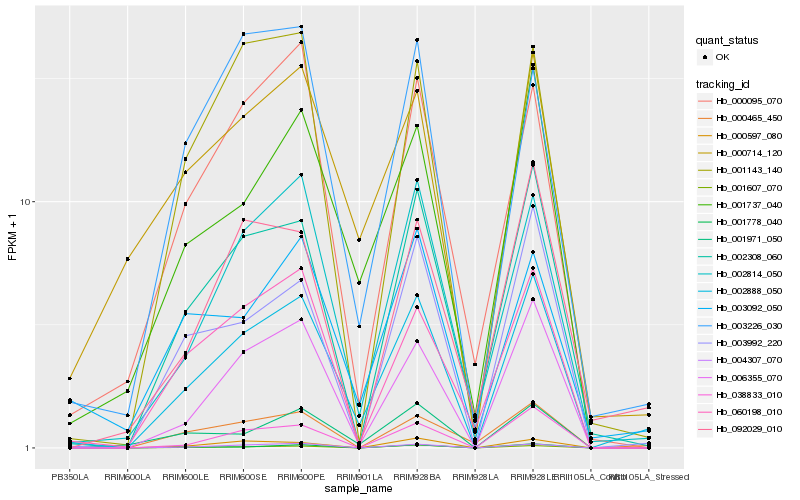
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002888_050 |
0.0 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002814_050 |
0.1389482554 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 3 |
Hb_006355_070 |
0.1410627892 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 4 |
Hb_002308_060 |
0.1487678988 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 5 |
Hb_001737_040 |
0.150986389 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 6 |
Hb_004307_070 |
0.1549978569 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_003992_220 |
0.1566283435 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 8 |
Hb_060198_010 |
0.1637995864 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 9 |
Hb_001607_070 |
0.1651213564 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 10 |
Hb_000465_450 |
0.1652077837 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 11 |
Hb_001778_040 |
0.1681979834 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |
| 12 |
Hb_001971_050 |
0.1687894096 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_038833_010 |
0.1695584147 |
- |
- |
polyprotein [Ananas comosus] |
| 14 |
Hb_003092_050 |
0.1707369537 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 15 |
Hb_000714_120 |
0.1737707563 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 16 |
Hb_001143_140 |
0.1745988114 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_003226_030 |
0.1750032367 |
- |
- |
sucrose synthase 5 [Hevea brasiliensis] |
| 18 |
Hb_000095_070 |
0.175091102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000597_080 |
0.1771456273 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_092029_010 |
0.177758478 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |