| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
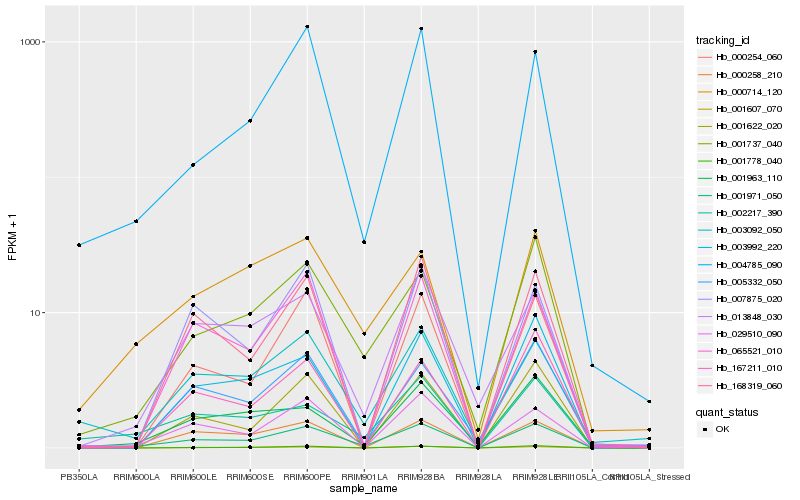
Hb_001971_050 |
0.0 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_001737_040 |
0.1325971158 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 3 |
Hb_029510_090 |
0.1353991611 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 4 |
Hb_001778_040 |
0.140931734 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |
| 5 |
Hb_013848_030 |
0.1448970617 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 6 |
Hb_000254_060 |
0.1524519727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004785_090 |
0.1534700936 |
- |
- |
hypothetical protein MIMGU_mgv1a018797mg [Erythranthe guttata] |
| 8 |
Hb_168319_060 |
0.1547752548 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Populus euphratica] |
| 9 |
Hb_065521_010 |
0.1556604473 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL5 [Jatropha curcas] |
| 10 |
Hb_001963_110 |
0.1571481514 |
- |
- |
- |
| 11 |
Hb_000258_210 |
0.15778309 |
- |
- |
prenyl-dependent CAAX protease, putative [Ricinus communis] |
| 12 |
Hb_003092_050 |
0.1578045444 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 13 |
Hb_001622_020 |
0.1581074946 |
- |
- |
- |
| 14 |
Hb_005332_050 |
0.1630957429 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 15 |
Hb_002217_390 |
0.1639678826 |
- |
- |
PREDICTED: galactokinase [Jatropha curcas] |
| 16 |
Hb_000714_120 |
0.1640719624 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 17 |
Hb_001607_070 |
0.1654226013 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 18 |
Hb_007875_020 |
0.1655243547 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 19 |
Hb_167211_010 |
0.1676787037 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_003992_220 |
0.1679754619 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |