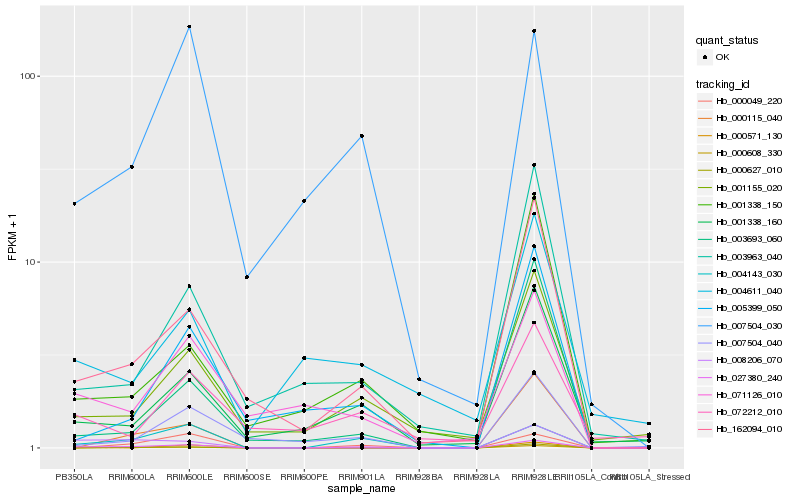
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027380_240 |
0.0 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004143_030 |
0.1915121648 |
- |
- |
PREDICTED: chromosome-associated kinesin KIF4B [Jatropha curcas] |
| 3 |
Hb_162094_010 |
0.2016645725 |
- |
- |
RNA polymerase alpha subunit [Hevea brasiliensis] |
| 4 |
Hb_001338_160 |
0.2332758299 |
- |
- |
NADH dehydrogenase subunit 5 [Hevea brasiliensis] |
| 5 |
Hb_001155_020 |
0.2379156447 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 2 [Theobroma cacao] |
| 6 |
Hb_008206_070 |
0.2548457754 |
- |
- |
hypothetical protein JCGZ_01495 [Jatropha curcas] |
| 7 |
Hb_000049_220 |
0.2562420692 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 8 |
Hb_007504_030 |
0.2566329328 |
- |
- |
Rpl14, partial (chloroplast) [Schistostemon retusum] |
| 9 |
Hb_071126_010 |
0.2581100507 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_007504_040 |
0.2672463369 |
- |
- |
hypothetical chloroplast RF2 [Fragaria iinumae] |
| 11 |
Hb_003963_040 |
0.273032311 |
- |
- |
ATP-dependent Clp protease proteolytic subunit [Hevea brasiliensis] |
| 12 |
Hb_005399_050 |
0.2778961955 |
- |
- |
DNA-directed RNA polymerase subunit beta [Glycine soja] |
| 13 |
Hb_001338_150 |
0.2815583891 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |
| 14 |
Hb_000608_330 |
0.2843783754 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 15 |
Hb_004611_040 |
0.2921730304 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigD, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_072212_010 |
0.2929915653 |
- |
- |
hypothetical chloroplast RF1 [Ficus sp. M. J. Moore 315] |
| 17 |
Hb_000115_040 |
0.2937132098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000571_130 |
0.2972324091 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_003693_060 |
0.2980843959 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 6 [Medicago truncatula] |
| 20 |
Hb_000627_010 |
0.2981419396 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |