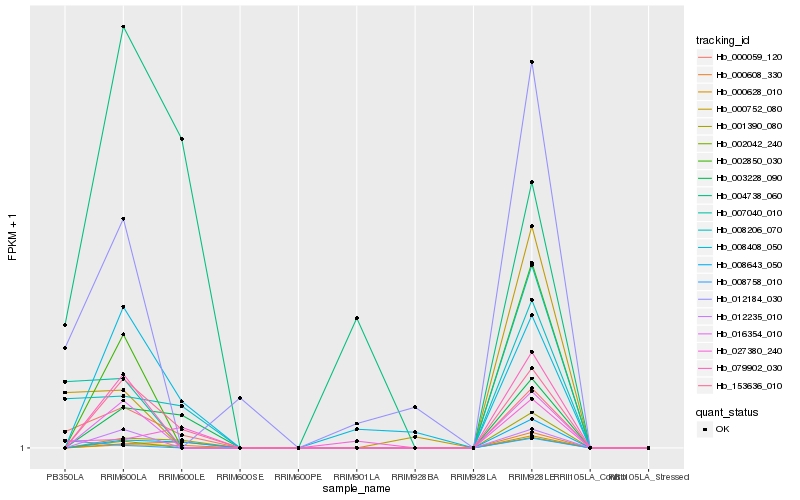
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_120 |
0.0 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 2 |
Hb_008206_070 |
0.1471882501 |
- |
- |
hypothetical protein JCGZ_01495 [Jatropha curcas] |
| 3 |
Hb_153636_010 |
0.2568837444 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 4 |
Hb_007040_010 |
0.2619248361 |
- |
- |
Uncharacterized protein TCM_024356 [Theobroma cacao] |
| 5 |
Hb_003228_090 |
0.2848842685 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105637258 [Jatropha curcas] |
| 6 |
Hb_008643_050 |
0.3007343968 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 7 |
Hb_001390_080 |
0.3010367768 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 8 |
Hb_000628_010 |
0.3027243176 |
- |
- |
PREDICTED: uncharacterized protein LOC105795966 [Gossypium raimondii] |
| 9 |
Hb_000752_080 |
0.3131000781 |
- |
- |
- |
| 10 |
Hb_012184_030 |
0.3261235969 |
- |
- |
- |
| 11 |
Hb_000608_330 |
0.3280644363 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 12 |
Hb_027380_240 |
0.3293269668 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004738_060 |
0.3346349386 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 14 |
Hb_008408_050 |
0.3372490557 |
- |
- |
- |
| 15 |
Hb_002042_240 |
0.3445519841 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_079902_030 |
0.345200572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002850_030 |
0.345949961 |
- |
- |
- |
| 18 |
Hb_016354_010 |
0.3470551643 |
- |
- |
- |
| 19 |
Hb_012235_010 |
0.3527081015 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 20 |
Hb_008758_010 |
0.3537495144 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |