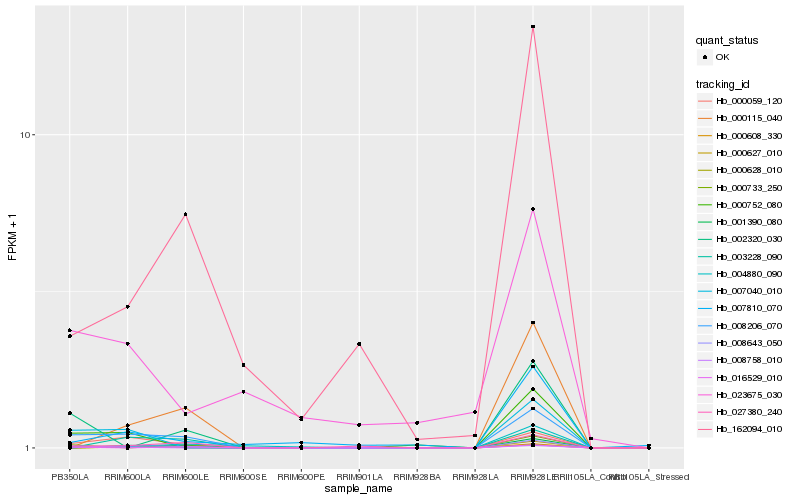
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008206_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_01495 [Jatropha curcas] |
| 2 |
Hb_000059_120 |
0.1471882501 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 3 |
Hb_027380_240 |
0.2548457754 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_007040_010 |
0.2678630584 |
- |
- |
Uncharacterized protein TCM_024356 [Theobroma cacao] |
| 5 |
Hb_001390_080 |
0.2939955722 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 6 |
Hb_000628_010 |
0.2947760332 |
- |
- |
PREDICTED: uncharacterized protein LOC105795966 [Gossypium raimondii] |
| 7 |
Hb_008643_050 |
0.2948785591 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 8 |
Hb_000752_080 |
0.2981292569 |
- |
- |
- |
| 9 |
Hb_000733_250 |
0.3025745885 |
- |
- |
serine/threonine protein phosphatase PP1 isozyme 1 [Loa loa] |
| 10 |
Hb_000608_330 |
0.3030624692 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 11 |
Hb_162094_010 |
0.3040826584 |
- |
- |
RNA polymerase alpha subunit [Hevea brasiliensis] |
| 12 |
Hb_002320_030 |
0.3090191769 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_016529_010 |
0.3102516078 |
- |
- |
polyprotein [Oryza australiensis] |
| 14 |
Hb_003228_090 |
0.3139872609 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105637258 [Jatropha curcas] |
| 15 |
Hb_000627_010 |
0.3172422784 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 16 |
Hb_008758_010 |
0.3191673849 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 17 |
Hb_000115_040 |
0.3212812431 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_023675_030 |
0.3340872703 |
- |
- |
- |
| 19 |
Hb_004880_090 |
0.3440480907 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 20 |
Hb_007810_070 |
0.3451310241 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |