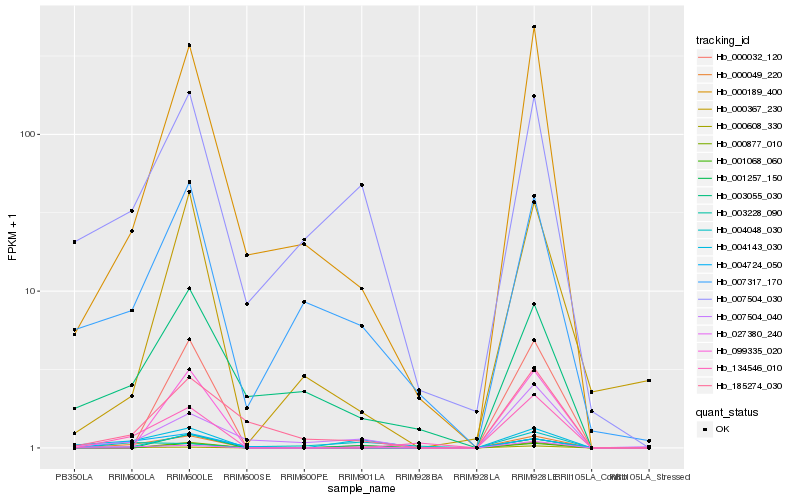
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000049_220 |
0.0 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 2 |
Hb_004143_030 |
0.1839658794 |
- |
- |
PREDICTED: chromosome-associated kinesin KIF4B [Jatropha curcas] |
| 3 |
Hb_007504_030 |
0.2420297603 |
- |
- |
Rpl14, partial (chloroplast) [Schistostemon retusum] |
| 4 |
Hb_134546_010 |
0.2437203934 |
- |
- |
PREDICTED: uncharacterized protein LOC105107372 [Populus euphratica] |
| 5 |
Hb_001257_150 |
0.253098703 |
- |
- |
- |
| 6 |
Hb_027380_240 |
0.2562420692 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000189_400 |
0.2565911181 |
- |
- |
hypothetical protein CathTA2_0066 [Caldalkalibacillus thermarum TA2.A1] |
| 8 |
Hb_000608_330 |
0.2653101508 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 9 |
Hb_004724_050 |
0.2710608195 |
transcription factor |
TF Family: bZIP |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007317_170 |
0.2718108975 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 11 |
Hb_000367_230 |
0.2907149462 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_004048_030 |
0.2920685288 |
- |
- |
hypothetical protein B456_009G371200, partial [Gossypium raimondii] |
| 13 |
Hb_185274_030 |
0.2929761436 |
- |
- |
hypothetical protein JCGZ_15170 [Jatropha curcas] |
| 14 |
Hb_003228_090 |
0.2939621265 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105637258 [Jatropha curcas] |
| 15 |
Hb_007504_040 |
0.3002820422 |
- |
- |
hypothetical chloroplast RF2 [Fragaria iinumae] |
| 16 |
Hb_003055_030 |
0.3064184277 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 17 |
Hb_000032_120 |
0.3096562046 |
- |
- |
Bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase [Glycine soja] |
| 18 |
Hb_001068_060 |
0.3096564946 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 19 |
Hb_099335_020 |
0.3096590983 |
- |
- |
hypothetical protein ARALYDRAFT_905869 [Arabidopsis lyrata subsp. lyrata] |
| 20 |
Hb_000877_010 |
0.3096705031 |
- |
- |
PREDICTED: F-box protein At3g07870-like [Jatropha curcas] |