| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
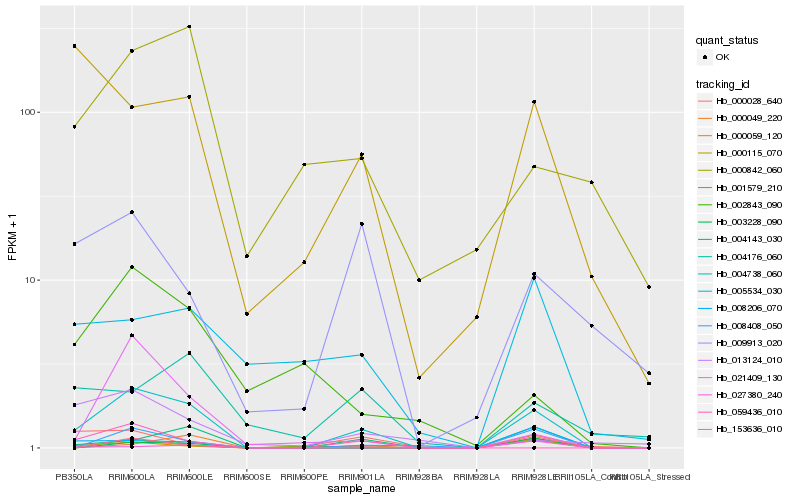
Hb_004738_060 |
0.0 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 2 |
Hb_004143_030 |
0.2894447223 |
- |
- |
PREDICTED: chromosome-associated kinesin KIF4B [Jatropha curcas] |
| 3 |
Hb_008408_050 |
0.3165378153 |
- |
- |
- |
| 4 |
Hb_000059_120 |
0.3346349386 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 5 |
Hb_001579_210 |
0.342906886 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_021409_130 |
0.3500254677 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like isoform 1 [Glycine max] |
| 7 |
Hb_000842_060 |
0.3560415137 |
- |
- |
PREDICTED: uncharacterized protein LOC105636760 [Jatropha curcas] |
| 8 |
Hb_027380_240 |
0.3593457119 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005534_030 |
0.3616361892 |
- |
- |
hypothetical protein JCGZ_00732 [Jatropha curcas] |
| 10 |
Hb_004176_060 |
0.3626593407 |
- |
- |
- |
| 11 |
Hb_009913_020 |
0.3629267375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003228_090 |
0.3675912102 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105637258 [Jatropha curcas] |
| 13 |
Hb_000115_070 |
0.3685766767 |
- |
- |
hypothetical protein RCOM_0030010 [Ricinus communis] |
| 14 |
Hb_002843_090 |
0.3706087816 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000049_220 |
0.3730685724 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 16 |
Hb_013124_010 |
0.3755911172 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_059436_010 |
0.3771827539 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000028_640 |
0.3779025286 |
- |
- |
Auxin efflux carrier component, putative [Ricinus communis] |
| 19 |
Hb_153636_010 |
0.3838794451 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 20 |
Hb_008206_070 |
0.3874088071 |
- |
- |
hypothetical protein JCGZ_01495 [Jatropha curcas] |