| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
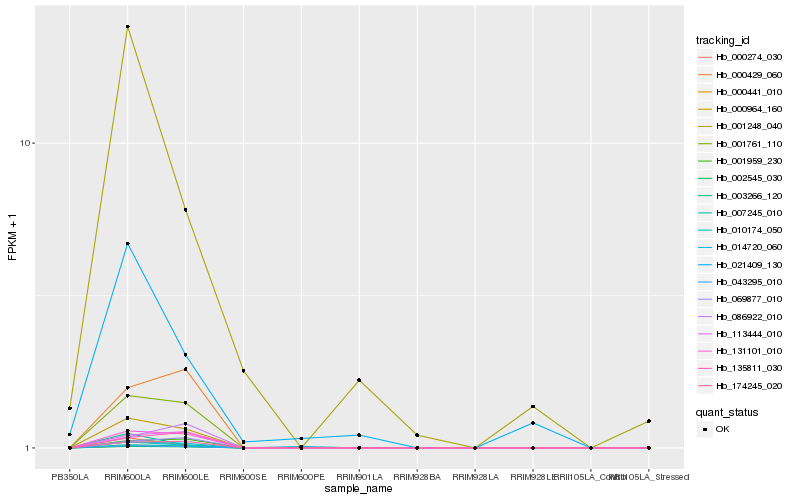
Hb_043295_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 2 |
Hb_007245_010 |
0.0001010675 |
- |
- |
PREDICTED: uncharacterized protein LOC104783698 [Camelina sativa] |
| 3 |
Hb_000964_160 |
0.0035346496 |
- |
- |
PREDICTED: uncharacterized protein LOC102617979 isoform X2 [Citrus sinensis] |
| 4 |
Hb_002545_030 |
0.0449483237 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_000441_010 |
0.0498757895 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 6 |
Hb_014720_060 |
0.0506000481 |
- |
- |
hypothetical protein JCGZ_11469 [Jatropha curcas] |
| 7 |
Hb_131101_010 |
0.0658940883 |
- |
- |
- |
| 8 |
Hb_001761_110 |
0.0702656862 |
- |
- |
Contains similarity to Cf-2.2 gene gb |
| 9 |
Hb_174245_020 |
0.1323138197 |
- |
- |
- |
| 10 |
Hb_001959_230 |
0.1527531358 |
- |
- |
PREDICTED: uncharacterized protein LOC105638436 [Jatropha curcas] |
| 11 |
Hb_113444_010 |
0.1618948074 |
- |
- |
- |
| 12 |
Hb_000274_030 |
0.1687145283 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 13 |
Hb_000429_060 |
0.1798276368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_069877_010 |
0.1969871585 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 15 |
Hb_003266_120 |
0.2010725069 |
- |
- |
PREDICTED: uncharacterized protein LOC105650228 [Jatropha curcas] |
| 16 |
Hb_135811_030 |
0.2015675701 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 17 |
Hb_021409_130 |
0.2714518286 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like isoform 1 [Glycine max] |
| 18 |
Hb_001248_040 |
0.2747203907 |
- |
- |
- |
| 19 |
Hb_010174_050 |
0.2810491923 |
- |
- |
PREDICTED: probable pectate lyase 5 [Populus euphratica] |
| 20 |
Hb_086922_010 |
0.2821214487 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |