| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
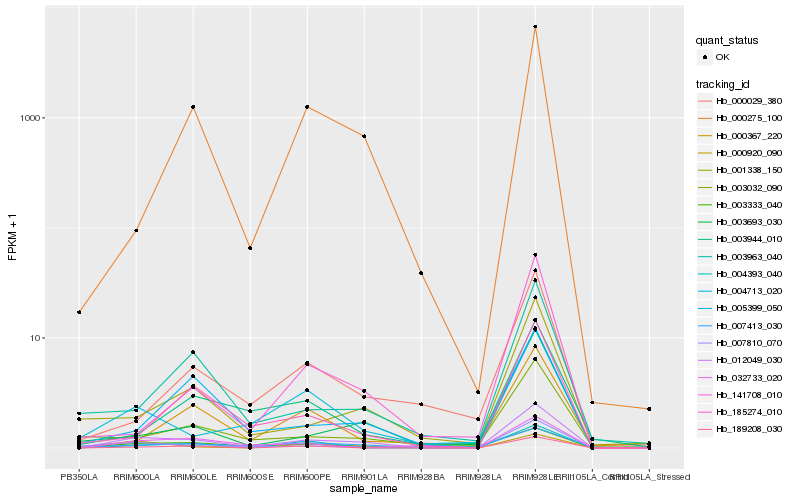
Hb_012049_030 |
0.0 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 2 |
Hb_000275_100 |
0.2123234768 |
- |
- |
BnaC09g26910D [Brassica napus] |
| 3 |
Hb_189208_030 |
0.2146574074 |
- |
- |
- |
| 4 |
Hb_004713_020 |
0.218657584 |
- |
- |
- |
| 5 |
Hb_003333_040 |
0.2279108827 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_004393_040 |
0.2284557712 |
transcription factor |
TF Family: RAV |
PREDICTED: AP2/ERF and B3 domain-containing transcription factor At1g50680-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_007413_030 |
0.2286960367 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_005399_050 |
0.2307636069 |
- |
- |
DNA-directed RNA polymerase subunit beta [Glycine soja] |
| 9 |
Hb_185274_010 |
0.2332635301 |
- |
- |
hypothetical protein ARALYDRAFT_330174 [Arabidopsis lyrata subsp. lyrata] |
| 10 |
Hb_000920_090 |
0.2333333138 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: vinorine synthase-like [Jatropha curcas] |
| 11 |
Hb_001338_150 |
0.2355305435 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |
| 12 |
Hb_007810_070 |
0.2364567041 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 13 |
Hb_003032_090 |
0.2398315271 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 14 |
Hb_000029_380 |
0.241297965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003963_040 |
0.2424302838 |
- |
- |
ATP-dependent Clp protease proteolytic subunit [Hevea brasiliensis] |
| 16 |
Hb_003693_030 |
0.2428126773 |
- |
- |
NADH dehydrogenase subunit 7 [Hevea brasiliensis] |
| 17 |
Hb_032733_020 |
0.2445849816 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X4 [Citrus sinensis] |
| 18 |
Hb_000367_220 |
0.2478863008 |
- |
- |
hypothetical protein RCOM_1470580 [Ricinus communis] |
| 19 |
Hb_003944_010 |
0.2490212634 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 20 |
Hb_141708_010 |
0.2495610104 |
- |
- |
hypothetical protein EUTSA_v10011731mg [Eutrema salsugineum] |