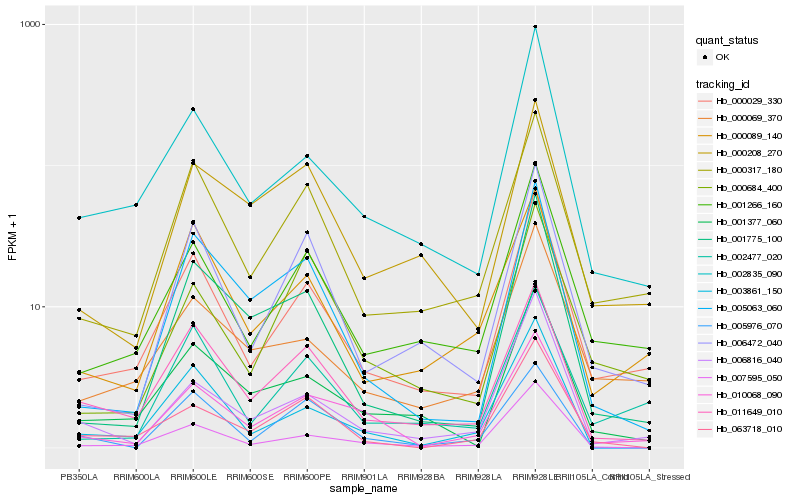
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010068_090 |
0.0 |
- |
- |
hypothetical protein JCGZ_00885 [Jatropha curcas] |
| 2 |
Hb_000029_330 |
0.1532419651 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000317_180 |
0.164467896 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_011649_010 |
0.1648126403 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 5 |
Hb_007595_050 |
0.1671222957 |
- |
- |
hypothetical chloroplast RF2 [Hevea brasiliensis] |
| 6 |
Hb_002835_090 |
0.1715312831 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 7 |
Hb_001266_160 |
0.174357318 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005063_060 |
0.1776849655 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 9 |
Hb_006816_040 |
0.1777483291 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910-like [Jatropha curcas] |
| 10 |
Hb_003861_150 |
0.178208662 |
- |
- |
hypothetical protein JCGZ_15167 [Jatropha curcas] |
| 11 |
Hb_000684_400 |
0.1841771465 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |
| 12 |
Hb_001377_060 |
0.1848525777 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |
| 13 |
Hb_063718_010 |
0.1865545912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000208_270 |
0.1866070862 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 15 |
Hb_001775_100 |
0.1867204485 |
- |
- |
PREDICTED: probable 2-carboxy-D-arabinitol-1-phosphatase [Jatropha curcas] |
| 16 |
Hb_000089_140 |
0.186983896 |
- |
- |
PREDICTED: uncharacterized protein LOC105636682 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000069_370 |
0.1880693171 |
- |
- |
- |
| 18 |
Hb_006472_040 |
0.1882859623 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005976_070 |
0.1902123626 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 20 |
Hb_002477_020 |
0.1903635488 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |