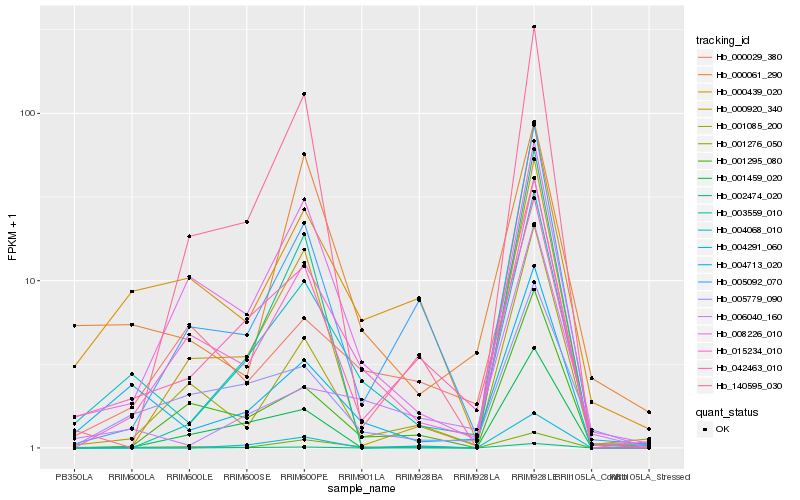
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004068_010 |
0.0 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 2 |
Hb_004713_020 |
0.1112448992 |
- |
- |
- |
| 3 |
Hb_015234_010 |
0.1707694545 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 4 |
Hb_042463_010 |
0.1848333094 |
- |
- |
cytochrome c oxidase subunit 2-1 (mitochondrion) (mitochondrion) [Brassica juncea var. tumida] |
| 5 |
Hb_001276_050 |
0.1902916321 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance-like protein 1O [Medicago truncatula] |
| 6 |
Hb_000439_020 |
0.1951271573 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3-like [Jatropha curcas] |
| 7 |
Hb_005779_090 |
0.2040333047 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 8 |
Hb_008226_010 |
0.2055625299 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 9 |
Hb_000920_340 |
0.2104350495 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_005092_070 |
0.2125169613 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 11 |
Hb_000061_290 |
0.2161144532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004291_060 |
0.2172151061 |
- |
- |
hypothetical protein F775_42457 [Aegilops tauschii] |
| 13 |
Hb_001085_200 |
0.2185797749 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 14 |
Hb_003559_010 |
0.2191613104 |
- |
- |
Peroxidase 27 precursor, putative [Ricinus communis] |
| 15 |
Hb_140595_030 |
0.2207927075 |
- |
- |
- |
| 16 |
Hb_000029_380 |
0.2247190227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006040_160 |
0.2256016575 |
- |
- |
ribosomal protein S12 (mitochondrion) [Hevea brasiliensis] |
| 18 |
Hb_001459_020 |
0.2267022053 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 19 |
Hb_001295_080 |
0.2287035605 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 20 |
Hb_002474_020 |
0.2291906659 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104585652 [Nelumbo nucifera] |