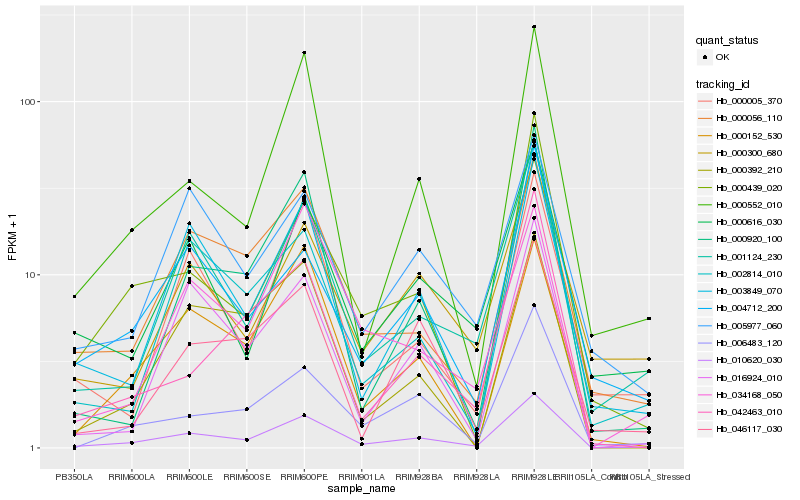
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010620_030 |
0.0 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 2 |
Hb_001124_230 |
0.130853851 |
- |
- |
PREDICTED: divinyl chlorophyllide a 8-vinyl-reductase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000552_010 |
0.1427453845 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 2 [Jatropha curcas] |
| 4 |
Hb_006483_120 |
0.1440633873 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_003849_070 |
0.1467558131 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 6 |
Hb_000056_110 |
0.1502871064 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 7 |
Hb_000439_020 |
0.1505842528 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3-like [Jatropha curcas] |
| 8 |
Hb_000152_530 |
0.1506904122 |
- |
- |
hypothetical protein SELMODRAFT_231485 [Selaginella moellendorffii] |
| 9 |
Hb_000616_030 |
0.15441929 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 10 |
Hb_000920_100 |
0.1566718667 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_016924_010 |
0.1570362593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004712_200 |
0.1620568425 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 13 |
Hb_000300_680 |
0.16617126 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 14 |
Hb_046117_030 |
0.1666797259 |
- |
- |
sialyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000005_370 |
0.1668018284 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634024 [Jatropha curcas] |
| 16 |
Hb_002814_010 |
0.1668193251 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 17 |
Hb_034168_050 |
0.1676687593 |
- |
- |
Photosystem I assembly protein Ycf3 [Medicago truncatula] |
| 18 |
Hb_000392_210 |
0.1687369017 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0004s04600g [Populus trichocarpa] |
| 19 |
Hb_005977_060 |
0.171902316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_042463_010 |
0.1719304883 |
- |
- |
cytochrome c oxidase subunit 2-1 (mitochondrion) (mitochondrion) [Brassica juncea var. tumida] |