| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
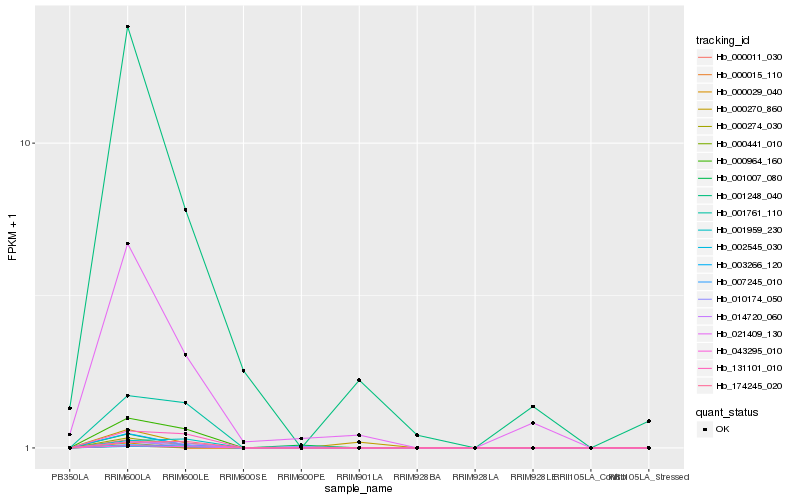
Hb_000274_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 2 |
Hb_003266_120 |
0.0330320307 |
- |
- |
PREDICTED: uncharacterized protein LOC105650228 [Jatropha curcas] |
| 3 |
Hb_043295_010 |
0.1687145283 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 4 |
Hb_007245_010 |
0.1688142688 |
- |
- |
PREDICTED: uncharacterized protein LOC104783698 [Camelina sativa] |
| 5 |
Hb_000964_160 |
0.1722020191 |
- |
- |
PREDICTED: uncharacterized protein LOC102617979 isoform X2 [Citrus sinensis] |
| 6 |
Hb_001248_040 |
0.2059402767 |
- |
- |
- |
| 7 |
Hb_002545_030 |
0.2129534207 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_000441_010 |
0.2177892834 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 9 |
Hb_014720_060 |
0.2184998584 |
- |
- |
hypothetical protein JCGZ_11469 [Jatropha curcas] |
| 10 |
Hb_021409_130 |
0.231728845 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like isoform 1 [Glycine max] |
| 11 |
Hb_131101_010 |
0.2334920395 |
- |
- |
- |
| 12 |
Hb_001761_110 |
0.2377729024 |
- |
- |
Contains similarity to Cf-2.2 gene gb |
| 13 |
Hb_001007_080 |
0.2744260355 |
- |
- |
PREDICTED: ribosome maturation protein SBDS [Jatropha curcas] |
| 14 |
Hb_010174_050 |
0.2979240655 |
- |
- |
PREDICTED: probable pectate lyase 5 [Populus euphratica] |
| 15 |
Hb_174245_020 |
0.2983316502 |
- |
- |
- |
| 16 |
Hb_001959_230 |
0.3182017574 |
- |
- |
PREDICTED: uncharacterized protein LOC105638436 [Jatropha curcas] |
| 17 |
Hb_000270_860 |
0.3215135489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000011_030 |
0.3252663637 |
- |
- |
hypothetical protein POPTR_0003s18920g [Populus trichocarpa] |
| 19 |
Hb_000015_110 |
0.3252663637 |
- |
- |
hypothetical protein MIMGU_mgv1a020753mg, partial [Erythranthe guttata] |
| 20 |
Hb_000029_040 |
0.3252663637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |