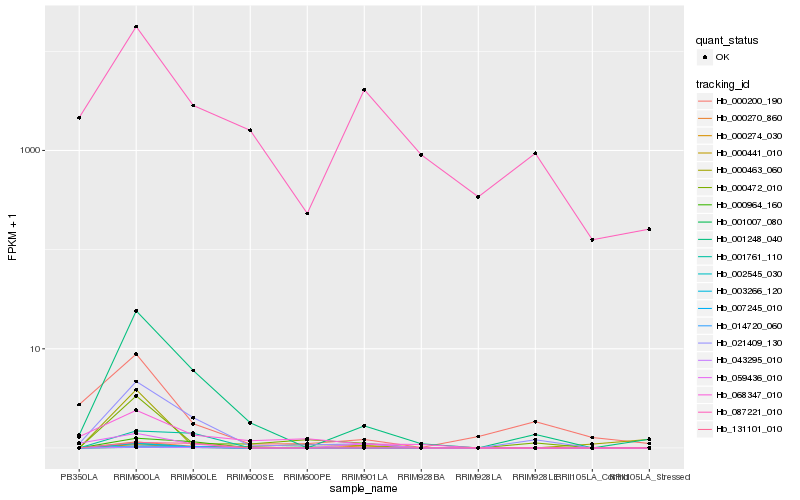
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001248_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_021409_130 |
0.153888857 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like isoform 1 [Glycine max] |
| 3 |
Hb_003266_120 |
0.2056079153 |
- |
- |
PREDICTED: uncharacterized protein LOC105650228 [Jatropha curcas] |
| 4 |
Hb_000274_030 |
0.2059402767 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 5 |
Hb_043295_010 |
0.2747203907 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 6 |
Hb_007245_010 |
0.2747849539 |
- |
- |
PREDICTED: uncharacterized protein LOC104783698 [Camelina sativa] |
| 7 |
Hb_000964_160 |
0.2769890978 |
- |
- |
PREDICTED: uncharacterized protein LOC102617979 isoform X2 [Citrus sinensis] |
| 8 |
Hb_000472_010 |
0.2967239111 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000270_860 |
0.2985639831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_087221_010 |
0.3008378217 |
- |
- |
hypothetical protein CISIN_1g033698mg [Citrus sinensis] |
| 11 |
Hb_002545_030 |
0.3050449438 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_000441_010 |
0.30854268 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 13 |
Hb_014720_060 |
0.3090593263 |
- |
- |
hypothetical protein JCGZ_11469 [Jatropha curcas] |
| 14 |
Hb_131101_010 |
0.3201133159 |
- |
- |
- |
| 15 |
Hb_001761_110 |
0.3233210183 |
- |
- |
Contains similarity to Cf-2.2 gene gb |
| 16 |
Hb_001007_080 |
0.334437388 |
- |
- |
PREDICTED: ribosome maturation protein SBDS [Jatropha curcas] |
| 17 |
Hb_000200_190 |
0.340967098 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 15 [Jatropha curcas] |
| 18 |
Hb_068347_010 |
0.3511108927 |
- |
- |
- |
| 19 |
Hb_000463_060 |
0.3515539363 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 20 |
Hb_059436_010 |
0.3584163194 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |