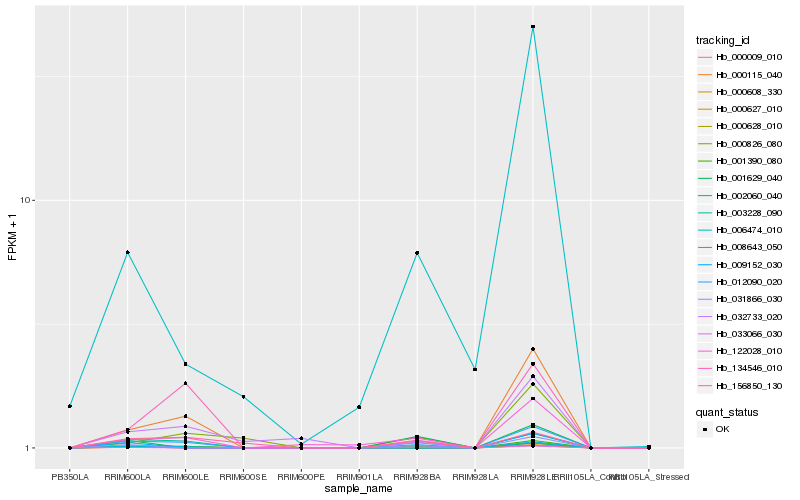
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012090_020 |
0.0 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 [Jatropha curcas] |
| 2 |
Hb_122028_010 |
0.2119777948 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 3 |
Hb_001390_080 |
0.2614070073 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 4 |
Hb_000628_010 |
0.2620426395 |
- |
- |
PREDICTED: uncharacterized protein LOC105795966 [Gossypium raimondii] |
| 5 |
Hb_008643_050 |
0.2622397865 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 6 |
Hb_134546_010 |
0.2670772071 |
- |
- |
PREDICTED: uncharacterized protein LOC105107372 [Populus euphratica] |
| 7 |
Hb_031866_030 |
0.2706002354 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis melo] |
| 8 |
Hb_002060_040 |
0.2718224985 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 9 |
Hb_033066_030 |
0.2719081628 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 10 |
Hb_009152_030 |
0.2723825296 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 11 |
Hb_000608_330 |
0.2728896503 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 12 |
Hb_032733_020 |
0.276563478 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X4 [Citrus sinensis] |
| 13 |
Hb_000826_080 |
0.2786446027 |
- |
- |
- |
| 14 |
Hb_156850_130 |
0.2800696494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000009_010 |
0.2832543386 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 16 |
Hb_000627_010 |
0.2838366794 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 17 |
Hb_000115_040 |
0.288236873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003228_090 |
0.2901693497 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105637258 [Jatropha curcas] |
| 19 |
Hb_006474_010 |
0.2919824571 |
- |
- |
hypothetical protein VITISV_032357 [Vitis vinifera] |
| 20 |
Hb_001629_040 |
0.2939319465 |
- |
- |
hypothetical protein POPTR_0001s41630g [Populus trichocarpa] |