| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
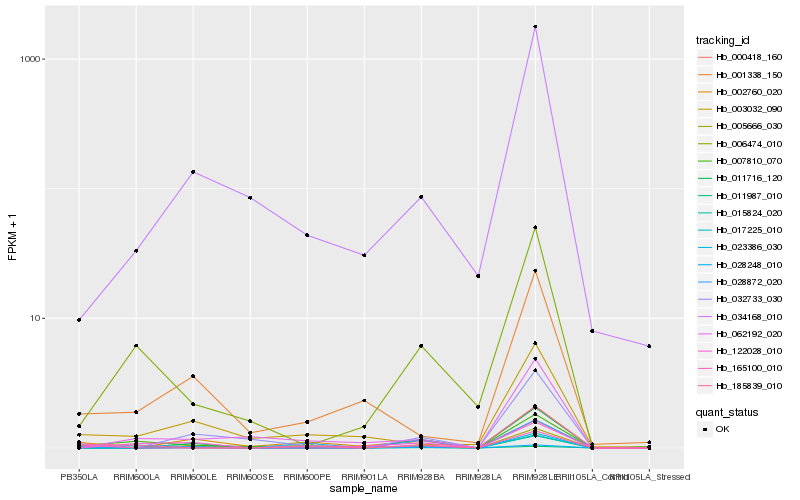
Hb_017225_010 |
0.0 |
- |
- |
PREDICTED: nodulation receptor kinase-like [Jatropha curcas] |
| 2 |
Hb_028248_010 |
0.1988834581 |
- |
- |
polyprotein [Oryza australiensis] |
| 3 |
Hb_011716_120 |
0.2002275736 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 4 |
Hb_185839_010 |
0.2123174233 |
- |
- |
- |
| 5 |
Hb_006474_010 |
0.2160883775 |
- |
- |
hypothetical protein VITISV_032357 [Vitis vinifera] |
| 6 |
Hb_002760_020 |
0.2281649889 |
- |
- |
PREDICTED: uncharacterized protein LOC105851424 [Cicer arietinum] |
| 7 |
Hb_122028_010 |
0.2302220174 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 8 |
Hb_001338_150 |
0.2483923726 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |
| 9 |
Hb_007810_070 |
0.2490752125 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 10 |
Hb_000418_160 |
0.2511157651 |
- |
- |
- |
| 11 |
Hb_034168_010 |
0.2546099461 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 12 |
Hb_165100_010 |
0.2561152048 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 13 |
Hb_003032_090 |
0.2580992114 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 14 |
Hb_032733_030 |
0.258466759 |
- |
- |
hypothetical protein CISIN_1g048657mg, partial [Citrus sinensis] |
| 15 |
Hb_028872_020 |
0.2595392813 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 16 |
Hb_011987_010 |
0.2600540935 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 17 |
Hb_023386_030 |
0.2603602111 |
- |
- |
- |
| 18 |
Hb_015824_020 |
0.2605554808 |
- |
- |
- |
| 19 |
Hb_062192_020 |
0.2637523784 |
- |
- |
cytochrome c biogenesis FN (mitochondrion) [Hevea brasiliensis] |
| 20 |
Hb_005666_030 |
0.2659353956 |
- |
- |
PREDICTED: uncharacterized protein LOC103489421 [Cucumis melo] |