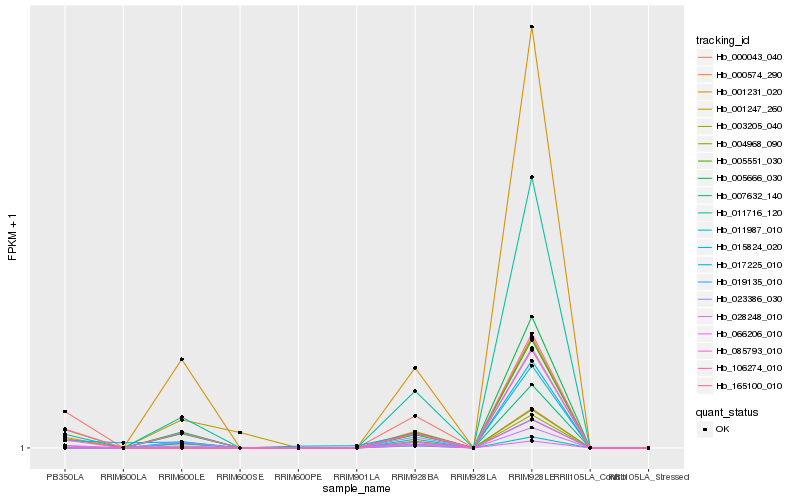
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028248_010 |
0.0 |
- |
- |
polyprotein [Oryza australiensis] |
| 2 |
Hb_011716_120 |
0.1159370857 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 3 |
Hb_017225_010 |
0.1988834581 |
- |
- |
PREDICTED: nodulation receptor kinase-like [Jatropha curcas] |
| 4 |
Hb_165100_010 |
0.203248929 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 5 |
Hb_011987_010 |
0.2126197268 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 6 |
Hb_085793_010 |
0.2184673312 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 7 |
Hb_003205_040 |
0.2190509486 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 8 |
Hb_106274_010 |
0.222290622 |
- |
- |
hypothetical protein VITISV_008825 [Vitis vinifera] |
| 9 |
Hb_001231_020 |
0.2255462882 |
- |
- |
- |
| 10 |
Hb_005551_030 |
0.2262676134 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 11 |
Hb_005666_030 |
0.2276370687 |
- |
- |
PREDICTED: uncharacterized protein LOC103489421 [Cucumis melo] |
| 12 |
Hb_000043_040 |
0.2281394175 |
- |
- |
CTV.19 [Citrus trifoliata] |
| 13 |
Hb_004968_090 |
0.2298456554 |
- |
- |
PREDICTED: uncharacterized protein LOC104882781 [Vitis vinifera] |
| 14 |
Hb_066206_010 |
0.2326097094 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_019135_010 |
0.2350701719 |
- |
- |
- |
| 16 |
Hb_015824_020 |
0.235137622 |
- |
- |
- |
| 17 |
Hb_023386_030 |
0.2354198156 |
- |
- |
- |
| 18 |
Hb_007632_140 |
0.2368224758 |
- |
- |
- |
| 19 |
Hb_000574_290 |
0.2384139943 |
- |
- |
PREDICTED: uncharacterized protein LOC105629978 [Jatropha curcas] |
| 20 |
Hb_001247_260 |
0.2385748597 |
- |
- |
- |