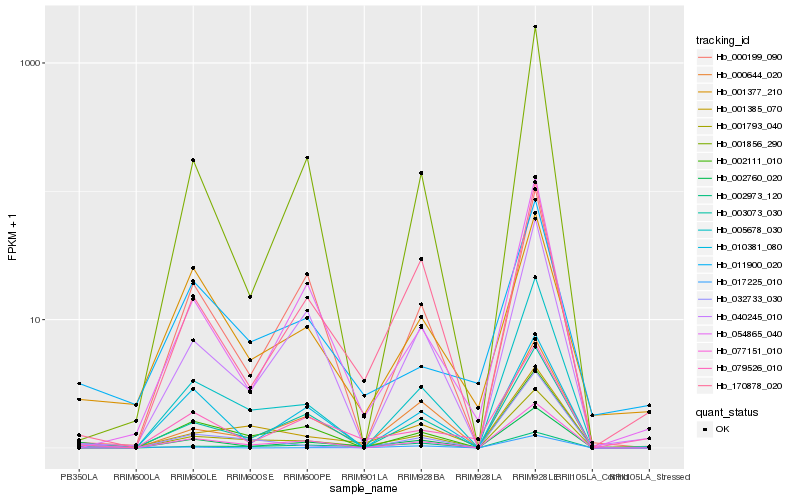
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002760_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105851424 [Cicer arietinum] |
| 2 |
Hb_001793_040 |
0.1790333712 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 3 |
Hb_000644_020 |
0.1927383344 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 4 |
Hb_170878_020 |
0.1973066456 |
- |
- |
- |
| 5 |
Hb_002973_120 |
0.2077558685 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 6 |
Hb_077151_010 |
0.2112724979 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 7 |
Hb_001385_070 |
0.2147985213 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 8 |
Hb_000199_090 |
0.2169003005 |
- |
- |
Tetrapyrrole-binding family protein [Populus trichocarpa] |
| 9 |
Hb_040245_010 |
0.2180435543 |
- |
- |
hypothetical protein CISIN_1g039575mg, partial [Citrus sinensis] |
| 10 |
Hb_003073_030 |
0.2190237012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_079526_010 |
0.2200246277 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG8 [Populus euphratica] |
| 12 |
Hb_010381_080 |
0.2203127933 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_032733_030 |
0.2211141592 |
- |
- |
hypothetical protein CISIN_1g048657mg, partial [Citrus sinensis] |
| 14 |
Hb_001377_210 |
0.2211538279 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 15 |
Hb_005678_030 |
0.2213287152 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 16 |
Hb_002111_010 |
0.2263381515 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 17 |
Hb_054865_040 |
0.2267384451 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 18 |
Hb_001856_290 |
0.2274922076 |
- |
- |
hypothetical protein JCGZ_16255 [Jatropha curcas] |
| 19 |
Hb_017225_010 |
0.2281649889 |
- |
- |
PREDICTED: nodulation receptor kinase-like [Jatropha curcas] |
| 20 |
Hb_011900_020 |
0.228191875 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |