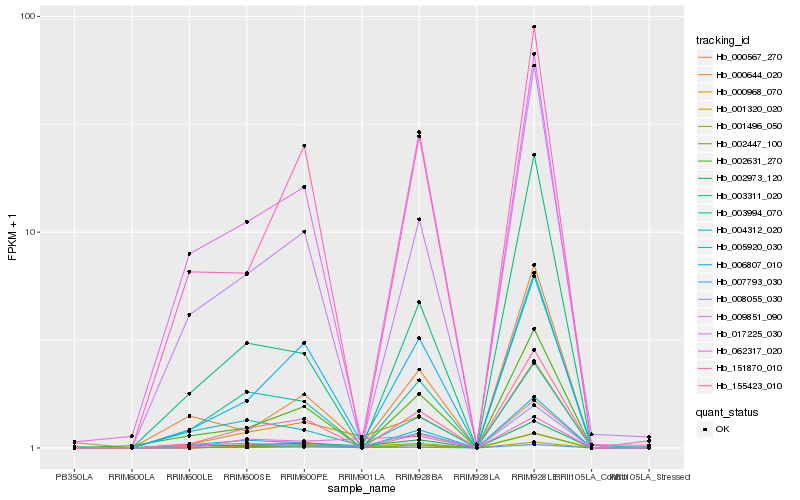
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000968_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_001496_050 |
0.1214083242 |
- |
- |
yth domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_007793_030 |
0.1229754039 |
- |
- |
OSJNBa0074B10.6 [Oryza sativa Japonica Group] |
| 4 |
Hb_001320_020 |
0.1599194878 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 5 |
Hb_155423_010 |
0.1601170128 |
- |
- |
- |
| 6 |
Hb_006807_010 |
0.1613959498 |
- |
- |
hypothetical protein JCGZ_20881 [Jatropha curcas] |
| 7 |
Hb_062317_020 |
0.1740006973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002631_270 |
0.1779248235 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s09930g [Populus trichocarpa] |
| 9 |
Hb_002973_120 |
0.180476987 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 10 |
Hb_009851_090 |
0.181331756 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 11 |
Hb_005920_030 |
0.1830328551 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 12 |
Hb_151870_010 |
0.1871485361 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_003994_070 |
0.1879842845 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 14 |
Hb_002447_100 |
0.1884924188 |
- |
- |
hypothetical protein POPTR_0009s06260g [Populus trichocarpa] |
| 15 |
Hb_004312_020 |
0.1893074708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_008055_030 |
0.1937821682 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 17 |
Hb_017225_030 |
0.1962088566 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 18 |
Hb_000567_270 |
0.1996500383 |
- |
- |
hypothetical protein L484_017162 [Morus notabilis] |
| 19 |
Hb_000644_020 |
0.200545676 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 20 |
Hb_003311_020 |
0.2027840796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |