| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
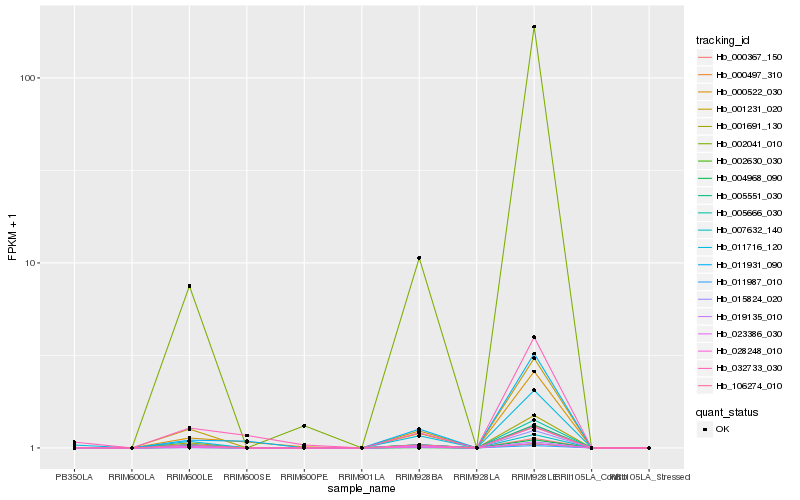
Hb_011716_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 2 |
Hb_028248_010 |
0.1159370857 |
- |
- |
polyprotein [Oryza australiensis] |
| 3 |
Hb_011987_010 |
0.1217228041 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 4 |
Hb_005666_030 |
0.1343709465 |
- |
- |
PREDICTED: uncharacterized protein LOC103489421 [Cucumis melo] |
| 5 |
Hb_005551_030 |
0.1351617514 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 6 |
Hb_106274_010 |
0.1358741096 |
- |
- |
hypothetical protein VITISV_008825 [Vitis vinifera] |
| 7 |
Hb_001231_020 |
0.1371451197 |
- |
- |
- |
| 8 |
Hb_004968_090 |
0.1373864534 |
- |
- |
PREDICTED: uncharacterized protein LOC104882781 [Vitis vinifera] |
| 9 |
Hb_015824_020 |
0.1376704835 |
- |
- |
- |
| 10 |
Hb_023386_030 |
0.1380004392 |
- |
- |
- |
| 11 |
Hb_001691_130 |
0.155355119 |
- |
- |
PREDICTED: uncharacterized protein LOC104587431 [Nelumbo nucifera] |
| 12 |
Hb_019135_010 |
0.1705911864 |
- |
- |
- |
| 13 |
Hb_002630_030 |
0.1709520016 |
- |
- |
OSIGBa0132I10.1 [Oryza sativa Indica Group] |
| 14 |
Hb_000497_310 |
0.1741649572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007632_140 |
0.1756666371 |
- |
- |
- |
| 16 |
Hb_011931_090 |
0.1842558674 |
- |
- |
- |
| 17 |
Hb_000367_150 |
0.184793108 |
- |
- |
- |
| 18 |
Hb_002041_010 |
0.186297942 |
- |
- |
- |
| 19 |
Hb_000522_030 |
0.1925833708 |
- |
- |
hypothetical protein VITISV_006017 [Vitis vinifera] |
| 20 |
Hb_032733_030 |
0.1936833303 |
- |
- |
hypothetical protein CISIN_1g048657mg, partial [Citrus sinensis] |