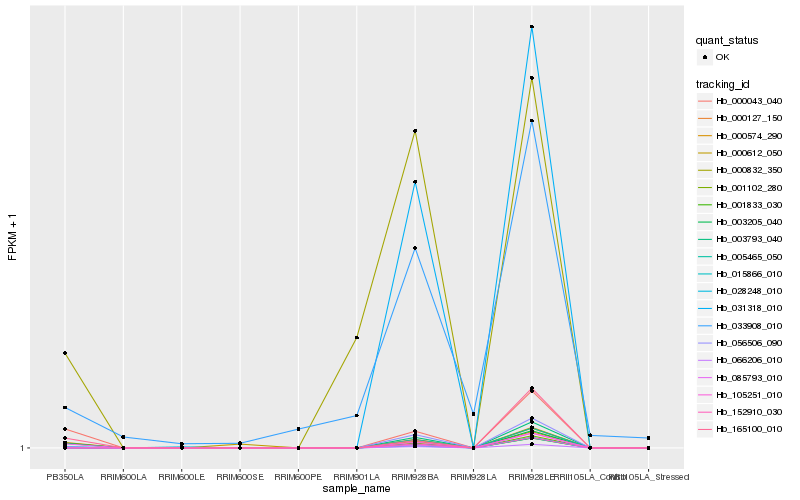
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_290 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629978 [Jatropha curcas] |
| 2 |
Hb_066206_010 |
0.0167712891 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_000043_040 |
0.0822816685 |
- |
- |
CTV.19 [Citrus trifoliata] |
| 4 |
Hb_003205_040 |
0.1185829697 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 5 |
Hb_085793_010 |
0.120216535 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 6 |
Hb_165100_010 |
0.2001167488 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 7 |
Hb_028248_010 |
0.2384139943 |
- |
- |
polyprotein [Oryza australiensis] |
| 8 |
Hb_000612_050 |
0.2505285868 |
- |
- |
hypothetical protein MIMGU_mgv1a020753mg, partial [Erythranthe guttata] |
| 9 |
Hb_000832_350 |
0.2522713288 |
- |
- |
Thylakoid lumenal 25.6 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_033908_010 |
0.2845514161 |
- |
- |
- |
| 11 |
Hb_005465_050 |
0.287285618 |
- |
- |
PREDICTED: uncharacterized protein LOC105635964 [Jatropha curcas] |
| 12 |
Hb_105251_010 |
0.2873452581 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_003793_040 |
0.2873771895 |
- |
- |
hypothetical protein CARUB_v10021783mg [Capsella rubella] |
| 14 |
Hb_001102_280 |
0.2874119694 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_031318_010 |
0.2874691845 |
- |
- |
- |
| 16 |
Hb_152910_030 |
0.287469271 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 17 |
Hb_056506_090 |
0.2874771609 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 18 |
Hb_000127_150 |
0.2874779647 |
- |
- |
PREDICTED: cytochrome P450 85A1-like [Jatropha curcas] |
| 19 |
Hb_015866_010 |
0.2874817776 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 20 |
Hb_001833_030 |
0.287487079 |
- |
- |
polyprotein [Oryza australiensis] |