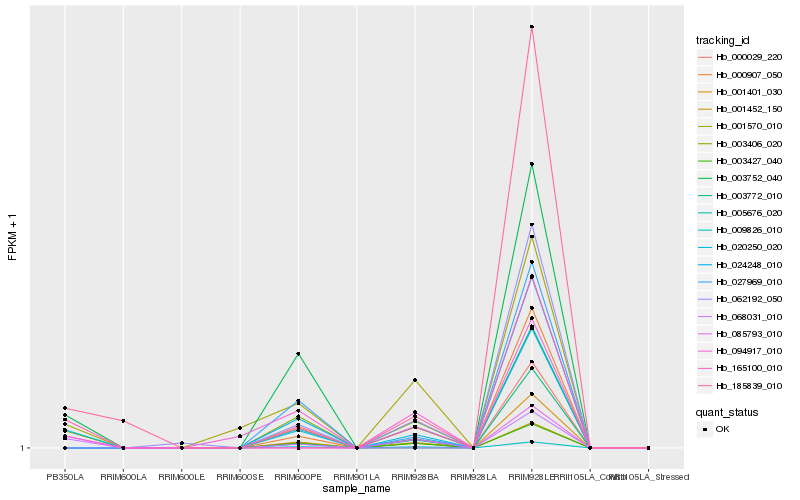
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020250_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 2 |
Hb_003406_020 |
0.1146753855 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 3 |
Hb_185839_010 |
0.210172737 |
- |
- |
- |
| 4 |
Hb_068031_010 |
0.2164148064 |
- |
- |
hypothetical protein VITISV_034229 [Vitis vinifera] |
| 5 |
Hb_001570_010 |
0.2171498265 |
- |
- |
- |
| 6 |
Hb_094917_010 |
0.2239673086 |
- |
- |
- |
| 7 |
Hb_165100_010 |
0.2241265499 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 8 |
Hb_024248_010 |
0.2299891163 |
- |
- |
PREDICTED: uncharacterized protein LOC105779006 [Gossypium raimondii] |
| 9 |
Hb_001401_030 |
0.231109165 |
- |
- |
PREDICTED: uncharacterized protein LOC105649622 [Jatropha curcas] |
| 10 |
Hb_003752_040 |
0.231177613 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 11 |
Hb_005676_020 |
0.234206202 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_1314900 [Ricinus communis] |
| 12 |
Hb_000907_050 |
0.2416599713 |
- |
- |
- |
| 13 |
Hb_000029_220 |
0.2432464561 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SLU7-A-like [Jatropha curcas] |
| 14 |
Hb_009826_010 |
0.2438336483 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_003427_040 |
0.2454187431 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 16 |
Hb_001452_150 |
0.2476655259 |
- |
- |
PREDICTED: uncharacterized protein LOC104879858 [Vitis vinifera] |
| 17 |
Hb_062192_050 |
0.2489324491 |
- |
- |
- |
| 18 |
Hb_027969_010 |
0.2527737653 |
- |
- |
hypothetical protein JCGZ_24556 [Jatropha curcas] |
| 19 |
Hb_003772_010 |
0.2530463337 |
- |
- |
polyprotein [Oryza australiensis] |
| 20 |
Hb_085793_010 |
0.2531683357 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |