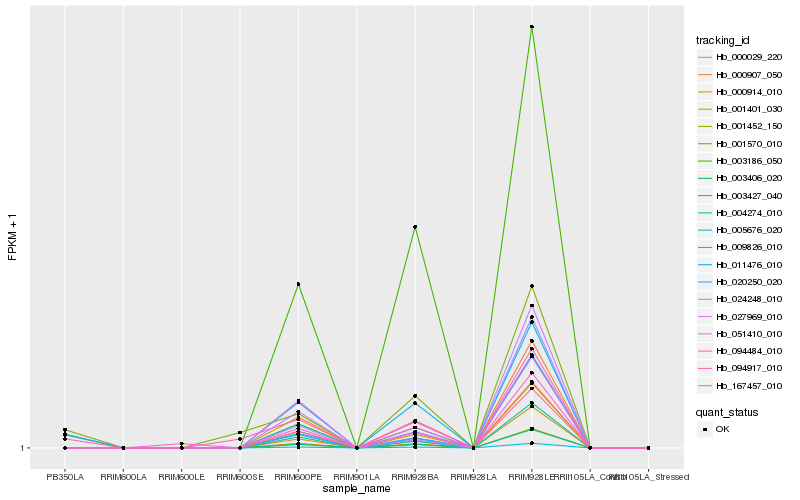
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003406_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 2 |
Hb_020250_020 |
0.1146753855 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 3 |
Hb_094917_010 |
0.1674245275 |
- |
- |
- |
| 4 |
Hb_001570_010 |
0.175376641 |
- |
- |
- |
| 5 |
Hb_004274_010 |
0.2063392397 |
- |
- |
PREDICTED: uncharacterized protein LOC104451199 [Eucalyptus grandis] |
| 6 |
Hb_001452_150 |
0.2075717127 |
- |
- |
PREDICTED: uncharacterized protein LOC104879858 [Vitis vinifera] |
| 7 |
Hb_003427_040 |
0.2091593305 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 8 |
Hb_009826_010 |
0.2096809704 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_011476_010 |
0.2203401078 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 10 |
Hb_000029_220 |
0.2256097335 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SLU7-A-like [Jatropha curcas] |
| 11 |
Hb_024248_010 |
0.2288675009 |
- |
- |
PREDICTED: uncharacterized protein LOC105779006 [Gossypium raimondii] |
| 12 |
Hb_003186_050 |
0.2326823582 |
- |
- |
- |
| 13 |
Hb_000914_010 |
0.2334651069 |
- |
- |
hypothetical protein POPTR_0025s00760g [Populus trichocarpa] |
| 14 |
Hb_167457_010 |
0.2359921975 |
- |
- |
hypothetical protein glysoja_045622, partial [Glycine soja] |
| 15 |
Hb_051410_010 |
0.2380786876 |
- |
- |
PREDICTED: uncharacterized protein LOC103401813 [Malus domestica] |
| 16 |
Hb_094484_010 |
0.2436758601 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_001401_030 |
0.2486570109 |
- |
- |
PREDICTED: uncharacterized protein LOC105649622 [Jatropha curcas] |
| 18 |
Hb_005676_020 |
0.2524787314 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_1314900 [Ricinus communis] |
| 19 |
Hb_000907_050 |
0.2566029474 |
- |
- |
- |
| 20 |
Hb_027969_010 |
0.2568670195 |
- |
- |
hypothetical protein JCGZ_24556 [Jatropha curcas] |